2JM6
 
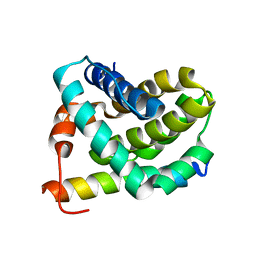 | | Solution structure of MCL-1 complexed with NOXAB | | Descriptor: | Myeloid cell leukemia-1 protein Mcl-1 homolog, Noxa | | Authors: | Czabotar, P.E, Lee, E.F, van Delft, M.F, Day, C.L, Smith, B.J, Huang, D.C.S, Fairlie, W.D, Hinds, M.G, Colman, P.M. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3ODA
 
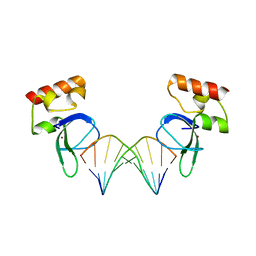 | | Human PARP-1 zinc finger 1 (Zn1) bound to DNA | | Descriptor: | 5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3', Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
1S4O
 
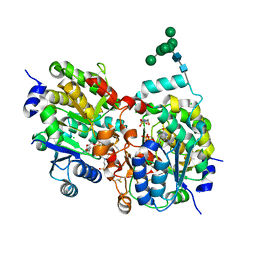 | | Crystal structure of yeast alpha1,2-mannosyltransferase Kre2p/Mnt1p: binary complex with GDP/Mn | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Lobsanov, Y.D, Romero, P.A, Sleno, B, Yu, B, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2004-01-16 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of Kre2p/Mnt1p: A YEAST {alpha}1,2-MANNOSYLTRANSFERASE INVOLVED IN MANNOPROTEIN BIOSYNTHESIS
J.Biol.Chem., 279, 2004
|
|
3OJV
 
 | | Crystal Structure of FGF1 complexed with the ectodomain of FGFR1c exhibiting an ordered ligand specificity-determining betaC'-betaE loop | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Basic fibroblast growth factor receptor 1, Heparin-binding growth factor 1 | | Authors: | Beenken, A, Mohammadi, M. | | Deposit date: | 2010-08-23 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Plasticity in Interactions of Fibroblast Growth Factor 1 (FGF1) N Terminus with FGF Receptors Underlies Promiscuity of FGF1.
J.Biol.Chem., 287, 2012
|
|
2QPU
 
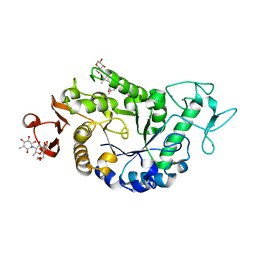 | | Sugar tongs mutant S378P in complex with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 1,5-anhydro-4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S,6R)-2,3,4,6-tetrahydroxy-5-methylcyclohexyl]amino}-alpha-D-glucopyranosyl)-D-glucitol, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Aghajari, N, Jensen, M.H, Tranier, S, Haser, R. | | Deposit date: | 2007-07-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 'pair of sugar tongs' site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity
Febs J., 274, 2007
|
|
2IIT
 
 | | Human dipeptidyl peptidase 4 in complex with a diazepan-2-one inhibitor | | Descriptor: | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-(2,2,2-TRIFLUOROETHYL)-1,4-DIAZEPAN-2-ONE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Biftu, T, Weber, A.E. | | Deposit date: | 2006-09-28 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | (3R)-4-[(3R)-3-Amino-4-(2,4,5-trifluorophenyl)butanoyl]-3-(2,2,2-trifluoroethyl)-1,4-diazepan-2-one, a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2EJN
 
 | | Structural characterization of the tetrameric form of the major cat allergen fel D 1 | | Descriptor: | CALCIUM ION, Major allergen I polypeptide chain 1, chain 2 | | Authors: | Kaiser, L, Velickovic, T.C, Badia-Martinez, D, Adedoyin, J, Thunberg, S, Hallen, D, Berndt, K, Gronlund, H, Gafvelin, G, van Hage, M, Achour, A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural characterization of the tetrameric form of the major cat allergen Fel d 1
J.Mol.Biol., 370, 2007
|
|
2J95
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5'-CHLORO-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-2,2'-BITHIOPHENE-5-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN, ... | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
7K51
 
 | | Mid-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K50
 
 | | Pre-translocation non-frameshifting(CCA-A) complex (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
2AES
 
 | | Crystal Structure of Human M340H-Beta1,4-Galactosyltransferase-I (M340H-B4Gal-T1) in Complex with GlcNAc-beta1,2-Man-alpha1,3-Man-beta-OR | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, ... | | Authors: | Ramasamy, V, Ramakrishnan, B, Boeggeman, E, Ratner, D.M, Seeberger, P.H, Qasba, P.K. | | Deposit date: | 2005-07-23 | | Release date: | 2005-10-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Preferences of beta1,4-Galactosyltransferase-I: Crystal Structures of Met340His Mutant of Human beta1,4-Galactosyltransferase-I with a Pentasaccharide and Trisaccharides of the N-Glycan Moiety
J.Mol.Biol., 353, 2005
|
|
7K52
 
 | | Near post-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
3B2D
 
 | | Crystal structure of human RP105/MD-1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD180 antigen, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2011-07-29 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Mouse and Human RP105/MD-1 Complexes Reveal Unique Dimer Organization of the Toll-Like Receptor Family.
J.Mol.Biol., 413, 2011
|
|
2KM2
 
 | | Galectin-1 dimer | | Descriptor: | Galectin-1 | | Authors: | Nesmelova, I.V, Ermakova, E, Daragan, V.A, Pang, M, Baum, L.G, Mayo, K.H. | | Deposit date: | 2009-07-16 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Lactose binding to galectin-1 modulates structural dynamics, increases conformational entropy, and occurs with apparent negative cooperativity.
J.Mol.Biol., 397, 2010
|
|
1TG7
 
 | | Native structure of beta-galactosidase from Penicillium sp. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rojas, A.L, Nagem, R.A.P, Neustroev, K.N, Arand, M, Adamska, M, Eneyskaya, E.V, Kulminskaya, A.A, Garratt, R.C, Golubev, A.M, Polikarpov, I. | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of beta-Galactosidase from Penicillium sp. and its Complex with Galactose
J.Mol.Biol., 343, 2004
|
|
2F6D
 
 | | Structure of the complex of a glucoamylase from Saccharomycopsis fibuligera with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Glucoamylase GLU1, PHOSPHATE ION, ... | | Authors: | Sevcik, J, Hostinova, E, Solovicova, A, Gasperik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2005-11-29 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the complex of a yeast glucoamylase with acarbose reveals the presence of a raw starch binding site on the catalytic domain.
Febs J., 273, 2006
|
|
2H54
 
 | |
2HE7
 
 | | FERM domain of EPB41L3 (DAL-1) | | Descriptor: | Band 4.1-like protein 3 | | Authors: | Hallberg, B.M, Busam, R.D, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Schiavone, L.H, Johansson, I, Hogbom, M, Karlberg, T, Kotenyova, T, Nilvebrandt, J, Norberg, P, Stenmark, P, Nordlund, P, Nilsson-ehle, P, Nyman, T, Ogg, D, Sagemark, J, Sundstrom, M, Uppenberg, J, Van den berg, S, Weigelt, J, Persson, C, Thorsell, A.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B).
J.Biol.Chem., 286, 2011
|
|
1V0Z
 
 | | Structure of Neuraminidase from English duck subtype N6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Rudino-Pinera, E, Tunnah, P, Crennell, S.J, Webster, R.G, Laver, W.G, Garman, E.F. | | Deposit date: | 2004-03-12 | | Release date: | 2006-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Crystal Structure of Influenza Type a Virus Neuraminidase of the N6 Subtype at 1.85 A Resolution
To be Published
|
|
2FJP
 
 | | Human dipeptidyl peptidase IV/CD26 in complex with an inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(4-{(1S,2S)-2-AMINO-1-[(DIMETHYLAMINO)CARBONYL]-3-[(3S)-3-FLUOROPYRROLIDIN-1-YL]-3-OXOPROPYL}PHENYL)-1H-[1,2,4]TRIAZOLO[1,5-A]PYRIDIN-4-IUM, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W. | | Deposit date: | 2006-01-03 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | (2S,3S)-3-Amino-4-(3,3-difluoropyrrolidin-1-yl)-N,N-dimethyl-4-oxo-2-(4-[1,2,4]triazolo[1,5-a]- pyridin-6-ylphenyl)butanamide: a selective alpha-amino amide dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
J.Med.Chem., 49, 2006
|
|
2ESC
 
 | | Crystal structure of a 40 KDa protective signalling protein from Bovine (SPC-40) at 2.1 A resolution | | Descriptor: | Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, J, Ethayathulla, A.S, Srivastav, D.B, Sharma, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a bovine secretory signalling glycoprotein (SPC-40) at 2.1 Angstrom resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2H4Y
 
 | |
1DTU
 
 | | BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE: A MUTANT Y89D/S146P COMPLEXED TO AN HEXASACCHARIDE INHIBITOR | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, PROTEIN (CYCLODEXTRIN GLYCOSYLTRANSFERASE), ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational design of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 to increase alpha-cyclodextrin production.
J.Mol.Biol., 296, 2000
|
|
2GYU
 
 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with HI-6 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Pang, Y.P, Boman, M, Artursson, E, Akfur, C, Lundberg, S. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: Structural basis for differences in the ability to reactivate tabun conjugates.
Biochem.Pharm., 72, 2006
|
|
2H51
 
 | |
