5DX0
 
 | |
5DWQ
 
 | |
8HE3
 
 | |
8HE0
 
 | |
8HLA
 
 | |
8HH0
 
 | |
3I8W
 
 | |
3OCQ
 
 | |
6Y5U
 
 | |
6Y56
 
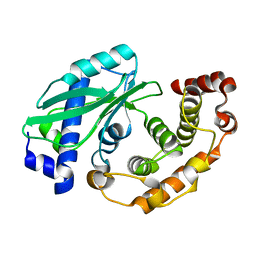 | |
9ASH
 
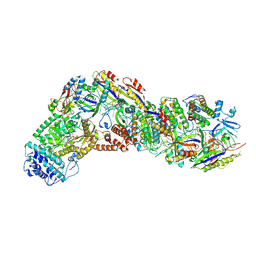 | |
1CSL
 
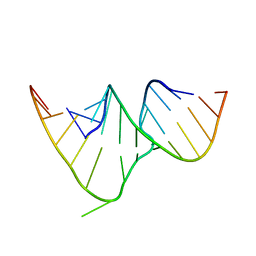 | | CRYSTAL STRUCTURE OF THE RRE HIGH AFFINITY SITE | | Descriptor: | 5'-R(*AP*AP*CP*GP*GP*GP*CP*GP*CP*AP*GP*AP*A)-3', 5'-R(*UP*CP*UP*GP*AP*CP*GP*GP*UP*AP*CP*GP*UP*UP*U)-3' | | Authors: | Ippolito, J.A, Steitz, T.A. | | Deposit date: | 1999-08-18 | | Release date: | 2000-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the HIV-1 RRE high affinity rev binding site at 1.6 A resolution.
J.Mol.Biol., 295, 2000
|
|
5UI6
 
 | | Solution NMR Structure of Lasso Peptide Acinetodin | | Descriptor: | Acinetodin | | Authors: | Bushin, L.B, Metelev, M, Severinov, K, Seyedsayamdost, M.R. | | Deposit date: | 2017-01-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae.
ACS Chem. Biol., 12, 2017
|
|
5UI7
 
 | | Solution NMR Structure of Lasso Peptide Klebsidin | | Descriptor: | Klebsidin | | Authors: | Bushin, L.B, Metelev, M, Severinov, K, Seyedsayamdost, M.R. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Acinetodin and Klebsidin, RNA Polymerase Targeting Lasso Peptides Produced by Human Isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae.
ACS Chem. Biol., 12, 2017
|
|
3B6E
 
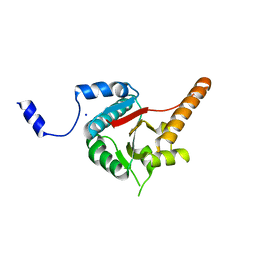 | | Crystal structure of human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, SODIUM ION | | Authors: | Karlberg, T, Welin, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human DECH-box RNA Helicase MDA5 (Melanoma differentiation-associated protein 5), DECH-domain.
To be Published
|
|
2X7K
 
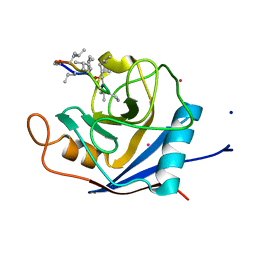 | | The crystal structure of PPIL1 in complex with cyclosporine A suggests a binding mode for SKIP | | Descriptor: | CADMIUM ION, CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE-LIKE 1, ... | | Authors: | Stegmann, C.M, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-23 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The Crystal Structure of Ppil1 Bound to Cyclosporine a Suggests a Binding Mode for a Linear Epitope of the Skip Protein.
Plos One, 5, 2010
|
|
1Q5K
 
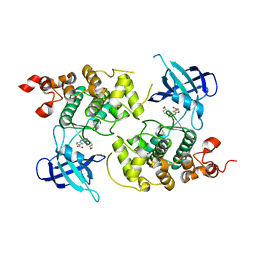 | | crystal structure of Glycogen synthase kinase 3 in complexed with inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-(4-METHOXYBENZYL)-N'-(5-NITRO-1,3-THIAZOL-2-YL)UREA | | Authors: | Bhat, R, Xue, Y, Berg, S, Hellberg, S, Ormo, M, Nilsson, Y, Radesater, A.C, Jerning, E, Markgren, P.O, Borgegard, T, Nylof, M, Gimenez-Cassina, A, Hernandez, F, Lucas, J.J, Diaz-Mido, J, Avila, J. | | Deposit date: | 2003-08-08 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418.
J.Biol.Chem., 278, 2003
|
|
3GX2
 
 | |
3GX6
 
 | |
3GX5
 
 | |
3GX7
 
 | |
3GX3
 
 | |
7GQT
 
 | | Crystal Structure of Werner helicase fragment 517-945 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, MAGNESIUM ION, ... | | Authors: | Classen, M, Benz, J, Brugger, D, Rudolph, M.G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase.
Nature, 629, 2024
|
|
7GQS
 
 | | Crystal Structure of Werner helicase fragment 517-945 in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ... | | Authors: | Classen, M, Benz, J, Brugger, D, Rudolph, M.G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase.
Nature, 629, 2024
|
|
8TOB
 
 | | Acinetobacter GP16 Type IV pilus | | Descriptor: | Fimbrial protein | | Authors: | Meng, R, Xing, Z, Zhang, J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
