1FUE
 
 | | FLAVODOXIN FROM HELICOBACTER PYLORI | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Freigang, J, Diederichs, K, Schaefer, K.P, Welte, W, Paul, R. | | Deposit date: | 2000-09-15 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori.
Protein Sci., 11, 2002
|
|
3B3P
 
 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-(3-chlorobenzyl)ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
1PX2
 
 | | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP (Form 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Synapsin I | | Authors: | Brautigam, C.A, Chelliah, Y, Deisenhofer, J. | | Deposit date: | 2003-07-02 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Tetramerization and ATP binding by a protein comprising the A, B, and C domains of rat synapsin I.
J.Biol.Chem., 279, 2004
|
|
7K1R
 
 | |
2EYX
 
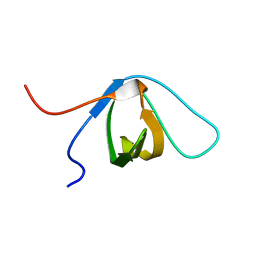 | |
2EYW
 
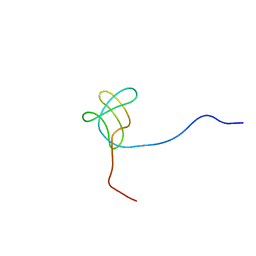 | |
2GQE
 
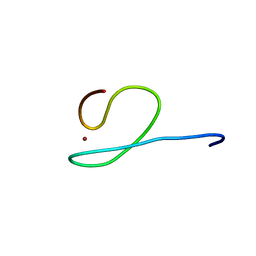 | | Molecular characterization of the Ran binding zinc finger domain | | Descriptor: | Nuclear pore complex protein Nup153, ZINC ION | | Authors: | Higa, M.M, Alam, S.L, Sundquist, W.I, Ullman, K.S. | | Deposit date: | 2006-04-20 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular Characterization of the Ran-binding Zinc Finger Domain of Nup153.
J.Biol.Chem., 282, 2007
|
|
2F5Z
 
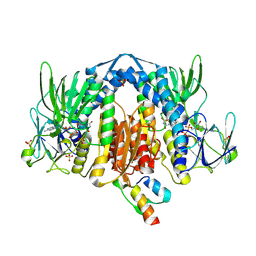 | | Crystal Structure of Human Dihydrolipoamide Dehydrogenase (E3) Complexed to the E3-Binding Domain of Human E3-Binding Protein | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, Pyruvate dehydrogenase protein X component, ... | | Authors: | Brautigam, C.A, Chuang, J.L, Wynn, R.M, Tomchick, D.R, Machius, M, Chuang, D.T. | | Deposit date: | 2005-11-28 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Insight into Interactions between Dihydrolipoamide Dehydrogenase (E3) and E3 Binding Protein of Human Pyruvate Dehydrogenase Complex.
Structure, 14, 2006
|
|
1IOZ
 
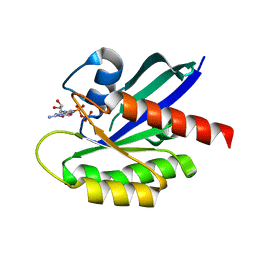 | | Crystal Structure of the C-HA-RAS Protein Prepared by the Cell-Free Synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Kigawa, T, Yamaguchi-Nunokawa, E, Kodama, K, Matsuda, T, Yabuki, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-04-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selenomethionine incorporation into a protein by cell-free synthesis
J.STRUCT.FUNCT.GENOM., 2, 2001
|
|
1KFS
 
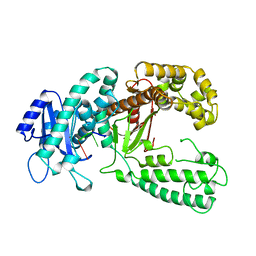 | | DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*AP*CP*G)-3'), MAGNESIUM ION, PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ... | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
2DVJ
 
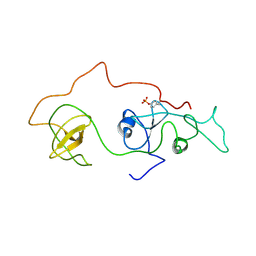 | | phosphorylated Crk-II | | Descriptor: | V-crk sarcoma virus CT10 oncogene homolog, isoform a | | Authors: | Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2006-07-31 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1X88
 
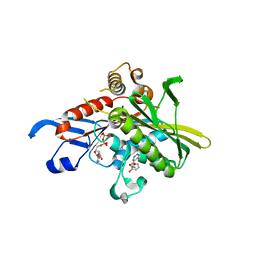 | | Human Eg5 motor domain bound to Mg-ADP and monastrol | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ETHYL 4-(3-HYDROXYPHENYL)-6-METHYL-2-THIOXO-1,2,3,4-TETRAHYDROPYRIMIDINE-5-CARBOXYLATE, Kinesin-like protein KIF11, ... | | Authors: | Maliga, Z, Mitchison, T.J. | | Deposit date: | 2004-08-17 | | Release date: | 2005-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Eg5 Inhibition by Monastrol
TO BE PUBLISHED
|
|
1VVJ
 
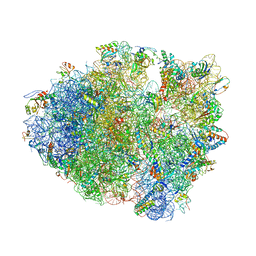 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 bound to Codon CCC-G on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-05-24 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.440001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3CM9
 
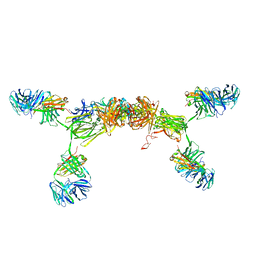 | | Solution Structure of Human SIgA2 | | Descriptor: | Immunoglobulin heavy chain, Immunoglobulin light chain, Secretory component | | Authors: | Bonner, A, Almogren, A, Furtado, P.B, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2008-03-21 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | The nonplanar secretory IgA2 and near planar secretory IgA1 solution structures rationalize their different mucosal immune responses.
J.Biol.Chem., 284, 2009
|
|
9QA6
 
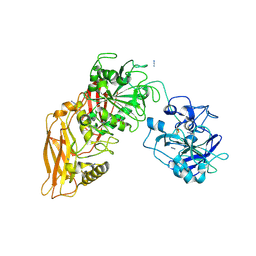 | | Cryo-EM structure of Thomasclavelia ramosa IgA peptidase (IgAse) active site mutant (S32-N876) | | Descriptor: | AZIDE ION, GLYCEROL, IgA protease, ... | | Authors: | Ramirez-Larrota, J.S, Eckhard, U, Guerra, P, Juyoux, P, Gomis-Ruth, F.X. | | Deposit date: | 2025-02-27 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Thomasclavelia ramosa IgA peptidase active site mutant - E540A (including NTD)
To Be Published
|
|
9I4Z
 
 | |
8P82
 
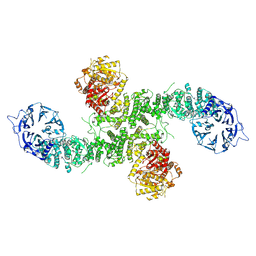 | | Cryo-EM structure of dimeric UBR5 | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-05-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
8P83
 
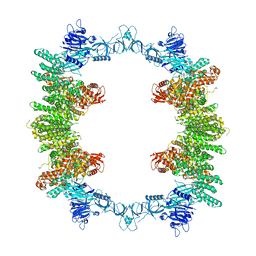 | | Cryo-EM structure of full-length human UBR5 (homotetramer) | | Descriptor: | E3 ubiquitin-protein ligase UBR5 | | Authors: | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-05-31 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
5DIG
 
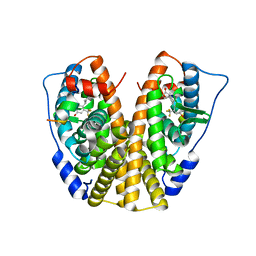 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a trifluoromethyl-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-5-[4-hydroxy-2-(trifluoromethyl)phenyl]-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-5-[4-hydroxy-2-(trifluoromethyl)phenyl]-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6FMJ
 
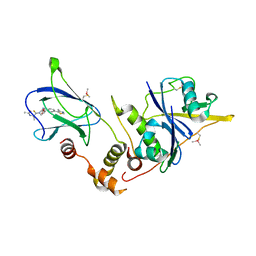 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-Acetamidopropanethioyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl) benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-acetamidopropanethioyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Soares, P, Lucas, X, Ciulli, A. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Thioamide substitution to probe the hydroxyproline recognition of VHL ligands.
Bioorg. Med. Chem., 26, 2018
|
|
6FMK
 
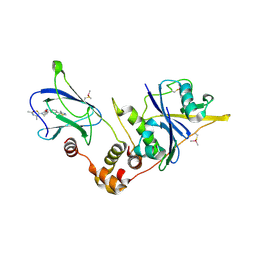 | | pVHL:EloB:EloC in complex with N-((S)-1-((2S,4R)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamothioyl) pyrrolidin-1-yl)-1-thioxopropan-2-yl)acetamide (ligand 4) | | Descriptor: | Elongin-B, Elongin-C, von Hippel-Lindau disease tumor suppressor, ... | | Authors: | Soares, P, Lucas, X, Ciulli, A. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Thioamide substitution to probe the hydroxyproline recognition of VHL ligands.
Bioorg. Med. Chem., 26, 2018
|
|
6FMI
 
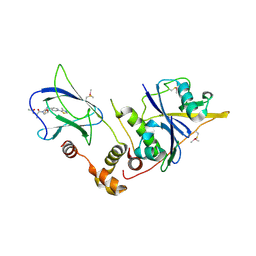 | | pVHL:EloB:EloC in complex with N-((S)-1-((2S,4R)-4-Hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamothioyl) pyrrolidin-1-yl)-1-oxopropan-2-yl)acetamide (ligand 2) | | Descriptor: | Elongin-B, Elongin-C, von Hippel-Lindau disease tumor suppressor, ... | | Authors: | Soares, P, Lucas, X, Ciulli, A. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thioamide substitution to probe the hydroxyproline recognition of VHL ligands.
Bioorg. Med. Chem., 26, 2018
|
|
5DIE
 
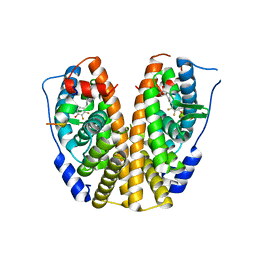 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a trifluoro-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-7a-methyl-5-(2,3,5-trifluoro-4-hydroxyphenyl)octahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-7a-methyl-5-(2,3,5-trifluoro-4-hydroxyphenyl)octahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DI7
 
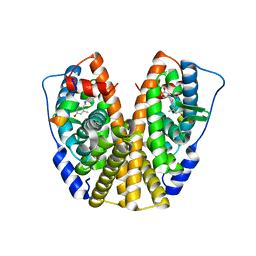 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with an methyl-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-5-(4-hydroxy-2-methylphenyl)-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-5-(4-hydroxy-2-methylphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DID
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a difluoro-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-5-(2,3-difluoro-4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-5-(2,3-difluoro-4-hydroxyphenyl)-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
