5MD4
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=6 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MDG
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=29, twist=0 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
2WGS
 
 | | Crystal structure of Mycobacterium Tuberculosis Glutamine Synthetase in complex with a purine analogue inhibitor. | | Descriptor: | 1-(3,4-dichlorobenzyl)-3,7-dimethyl-8-morpholin-4-yl-3,7-dihydro-1H-purine-2,6-dione, CHLORIDE ION, GLUTAMINE SYNTHETASE 1 | | Authors: | Nilsson, M.T, Krajewski, W.W, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2009-04-27 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterium Tuberculosis Glutamine Synthetase by Novel ATP-Competitive Inhibitors.
J.Mol.Biol., 393, 2009
|
|
6A5Z
 
 | | Crystal structure of human FXR/RXR-LBD heterodimer bound to HNC180 and 9cRA and SRC1 | | Descriptor: | (9cis)-retinoic acid, 2-[(1R,5S)-9-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-3-azabicyclo[3.3.1]nonan-3-yl]-1,3-benzothiazole-6-carboxylic acid, Bile acid receptor, ... | | Authors: | Wang, N, Liu, J. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Ligand binding and heterodimerization with retinoid X receptor alpha (RXR alpha ) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding
J. Biol. Chem., 293, 2018
|
|
2W8H
 
 | | Crystal structure of spin labeled Wza24-345. | | Descriptor: | CHLORIDE ION, PUTATIVE OUTER MEMBRANE LIPOPROTEIN WZA, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Hagelueken, G, Ingledew, W.J, Huang, H, Petrovic-Stojanovska, B, Whitfield, C, ElMkami, H, Schiemann, O, Naismith, J.H. | | Deposit date: | 2009-01-16 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Peldor Distance Fingerprinting of the Octameric Outer-Membrane Protein Wza from Escherichia Coli.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1YKJ
 
 | | A45G p-hydroxybenzoate hydroxylase with p-hydroxybenzoate bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, P-hydroxybenzoate hydroxylase, ... | | Authors: | Cole, L.J, Gatti, D.L, Entsch, B, Ballou, D.P. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Removal of a methyl group causes global changes in p-hydroxybenzoate hydroxylase.
Biochemistry, 44, 2005
|
|
1KEK
 
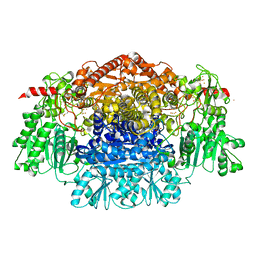 | | Crystal Structure of the Free Radical Intermediate of Pyruvate:Ferredoxin Oxidoreductase | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, CALCIUM ION, CARBON DIOXIDE, ... | | Authors: | Chabriere, E, Vernede, X, Guigliarelli, B, Charon, M.-H, Hatchikian, E.C, Fontecilla-Camps, J.C. | | Deposit date: | 2001-11-16 | | Release date: | 2001-12-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the free radical intermediate of pyruvate:ferredoxin oxidoreductase.
Science, 294, 2001
|
|
2VXS
 
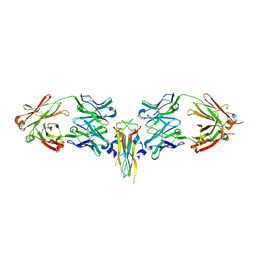 | | Structure of IL-17A in complex with a potent, fully human neutralising antibody | | Descriptor: | FAB FRAGMENT, INTERLEUKIN-17A, SULFATE ION | | Authors: | Gerhardt, S, Hargreaves, D, Pauptit, R.A, Davies, R.A, Russell, C, Welsh, F, Tuske, S.J, Coales, S.J, Hamuro, Y, Needham, M.R.C, Langham, C, Barker, W, Bell, P, Aziz, A, Smith, M.J, Dawson, S, Abbott, W.M. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure of Il-17A in Complex with a Potent, Fully Human Neutralising Antibody.
J.Mol.Biol., 394, 2009
|
|
1YE9
 
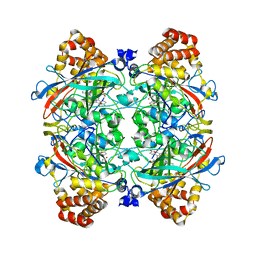 | | Crystal structure of proteolytically truncated catalase HPII from E. coli | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, catalase HPII | | Authors: | Loewen, P.C, Chelikani, P, Carpena, X, Fita, I, Perez-Luque, R, Donald, L.J, Switala, J, Duckworth, H.W. | | Deposit date: | 2004-12-28 | | Release date: | 2005-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of a Large Subunit Catalase Truncated by Proteolytic Cleavage(,)
Biochemistry, 44, 2005
|
|
2VV4
 
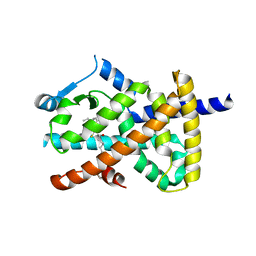 | | hPPARgamma Ligand binding domain in complex with 6-oxoOTE | | Descriptor: | (8E,10S,12Z)-10-hydroxy-6-oxooctadeca-8,12-dienoic acid, (8R,9Z,12Z)-8-hydroxy-6-oxooctadeca-9,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
5MD8
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=17, twist=12 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
2VXR
 
 | |
1KJ2
 
 | | Murine Alloreactive ScFv TCR-Peptide-MHC Class I Molecule Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Allogeneic H-2Kb MHC Class I Molecule, Beta-2 microglobulin, ... | | Authors: | Reiser, J.-B, Gregoire, C, Darnault, C, Mosser, T, Guimezanes, A, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Mazza, G, Malissen, B, Housset, D. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex.
Immunity, 16, 2002
|
|
5ZRF
 
 | | Crystal structure of human topoisomerase II beta in complex with 5-iodouridine-containing-DNA and etoposide in space group p21 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA topoisomerase 2-beta, ... | | Authors: | Chen, S.F, Wang, Y.R, Wu, C.C, Chan, N.L. | | Deposit date: | 2018-04-24 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the gating of DNA passage by the topoisomerase II DNA-gate.
Nat Commun, 9, 2018
|
|
1KJV
 
 | | TAP-B-associated rat MHC class I molecule | | Descriptor: | Mature alpha chain of major histocompatibility complex class I antigen (HEAVY CHAIN), SULFATE ION, beta-2-microglobulin, ... | | Authors: | Rudolph, M.G, Stevens, J, Speir, J.A, Trowsdale, J, Butcher, G.W, Joly, E, Wilson, I.A. | | Deposit date: | 2001-12-05 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of two rat MHC class Ia (RT1-A) molecules that are associated differentially
with peptide transporter alleles TAP-A and TAP-B.
J.Mol.Biol., 324, 2002
|
|
1YCI
 
 | |
2IHB
 
 | | Crystal structure of the heterodimeric complex of human RGS10 and activated Gi alpha 3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(k) subunit alpha, MAGNESIUM ION, ... | | Authors: | Soundararajan, M, Turnbull, A.P, Papagrigoriou, E, Pike, A.C.W, Bunkoczi, G, Ugochukwu, E, Gorrec, F, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1YKL
 
 | | Protocatechuate 3,4-Dioxygenase Y408C mutant bound to DHB | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
3GQ9
 
 | |
1YKK
 
 | | Protocatechuate 3,4-Dioxygenase Y408C Mutant | | Descriptor: | FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, Protocatechuate 3,4-dioxygenase beta chain | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1YG6
 
 | | ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Bewley, M.C, Graziano, V, Griffin, K, Flanagan, J.M. | | Deposit date: | 2005-01-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes.
J.Struct.Biol., 153, 2006
|
|
2P0D
 
 | | ArhGAP9 PH domain in complex with Ins(1,4,5)P3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Rho GTPase-activating protein 9 | | Authors: | Ceccarelli, D.F.J, Blasutig, I, Goudreault, M, Ruston, J, Pawson, T, Sicheri, F. | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Non-canonical Interaction of Phosphoinositides with Pleckstrin Homology Domains of Tiam1 and ArhGAP9.
J.Biol.Chem., 282, 2007
|
|
1YJ3
 
 | | Crystal structure analysis of product bound methionine aminopeptidase Type 1c from Mycobacterium Tuberculosis | | Descriptor: | BETA-MERCAPTOETHANOL, COBALT (II) ION, METHIONINE, ... | | Authors: | Addlagatta, A, Quillin, M.L, Omotoso, O, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-01-13 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of an SH3-binding motif in a new class of methionine aminopeptidases from Mycobacterium tuberculosis suggests a mode of interaction with the ribosome.
Biochemistry, 44, 2005
|
|
1YKM
 
 | | Protocatechuate 3,4-Dioxygenase Y408E mutant | | Descriptor: | FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, Protocatechuate 3,4-dioxygenase beta chain | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
2IQ1
 
 | | Crystal structure of human PPM1K | | Descriptor: | MAGNESIUM ION, Protein phosphatase 2C kappa, PPM1K | | Authors: | Bonanno, J.B, Freeman, J, Russell, M, Bain, K.T, Adams, J, Pelletier, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
