3OM3
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4BXS
 
 | | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lechtenberg, B.C, Murray-Rust, T.A, Johnson, D.J.D, Adams, T.E, Krishnaswamy, S, Camire, R.M, Huntington, J.A. | | Deposit date: | 2013-07-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal Structure of the Prothrombinase Complex from the Venom of Pseudonaja Textilis.
Blood, 122, 2013
|
|
5WJ1
 
 | | Crystal structure of Arabidopsis thaliana acetohydroxyacid synthase in complex with a triazolopyrimidine herbicide, penoxsulam | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2-(2,2-difluoroethoxy)-N-(5,8-dimethoxy[1,2,4]triazolo[1,5-c]pyrimidin-2-yl)-6-(trifluoromethyl)benzenesulfonamide, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2017-07-21 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structural insights into the mechanism of inhibition of AHAS by herbicides.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4BLW
 
 | | Crystal structure of Escherichia coli 23S rRNA (A2030-N6)- methyltransferase RlmJ in complex with S-adenosylhomocysteine (AdoHcy) and Adenosine monophosphate (AMP) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Punekar, A.S, Liljeruhm, J, Shepherd, T.R, Forster, A.C, Selmer, M. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Insights Into the Molecular Mechanism of Rrna M6A Methyltransferase Rlmj.
Nucleic Acids Res., 41, 2013
|
|
4AH2
 
 | | HLA-DR1 with covalently linked CLIP106-120 in canonical orientation | | Descriptor: | GLYCEROL, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN GAMMA CHAIN, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN,DRB1-1 BETA CHAIN, ... | | Authors: | Schlundt, A, Guenther, S, Sticht, J, Wieczorek, M, Roske, Y, Heinemann, U, Freund, C. | | Deposit date: | 2012-02-03 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Peptide Linkage to the Alpha-Subunit of Mhcii Creates a Stably Inverted Antigen Presentation Complex.
J.Mol.Biol., 423, 2012
|
|
3HAQ
 
 | | Crystal structure of bacteriorhodopsin mutant I148A crystallized from bicelles | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, DECANE, ... | | Authors: | Joh, N.H, Yang, D, Bowie, J.U. | | Deposit date: | 2009-05-02 | | Release date: | 2009-09-22 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Similar energetic contributions of packing in the core of membrane and water-soluble proteins.
J.Am.Chem.Soc., 131, 2009
|
|
3STL
 
 | | KcsA potassium channel mutant Y82C with Cadmium bound | | Descriptor: | CADMIUM ION, POTASSIUM ION, Voltage-gated potassium channel, ... | | Authors: | Raghuraman, H, Cordero-Morales, J, Jogini, V, Perozo, E. | | Deposit date: | 2011-07-11 | | Release date: | 2012-04-18 | | Last modified: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Cd(2+) Coordination during Slow Inactivation in Potassium Channels.
Structure, 20, 2012
|
|
4A6D
 
 | | Crystal structure of human N-acetylserotonin methyltransferase (ASMT) in complex with SAM | | Descriptor: | GLYCEROL, HYDROXYINDOLE O-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE, ... | | Authors: | Legrand, P, Haouz, A, Shepard, W. | | Deposit date: | 2011-11-01 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure and Functional Mapping of Human Asmt, the Last Enzyme of the Melatonin Synthesis Pathway.
J.Pineal Res., 54, 2013
|
|
4EQV
 
 | |
3SVP
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R)-4-(2-((2,2-Difluoro-2-(3-chloro-5-fluorophenyl)ethyl)amino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(2-{[2-(3-chloro-5-fluorophenyl)-2,2-difluoroethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2011-07-12 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Improved Synthesis of Chiral Pyrrolidine Inhibitors and Their Binding Properties to Neuronal Nitric Oxide Synthase.
J.Med.Chem., 54, 2011
|
|
5WOG
 
 | |
4AH4
 
 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) in complex with BGA Oligosaccharide. | | Descriptor: | CHLORIDE ION, FUCOSE-SPECIFIC LECTIN FLEA, alpha-L-fucopyranose, ... | | Authors: | Houser, J, Komarek, J, Kostlanova, N, Lahmann, M, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2012-02-03 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5WS3
 
 | | Crystal structures of human orexin 2 receptor bound to the selective antagonist EMPA determined by serial femtosecond crystallography at SACLA | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Kimura, K, Nakane, T, Yamashita, K, Wang, J, Fujiwara, T, Yamanaka, Y, Im, D, Tsujimoto, H, Sasanuma, M, Horita, S, Hirokawa, T, Nango, E, Tono, K, Kameshima, T, Hatsui, T, Joti, Y, Yabashi, M, Shimamoto, K, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA.
Structure, 26, 2018
|
|
3PAW
 
 | |
4AKL
 
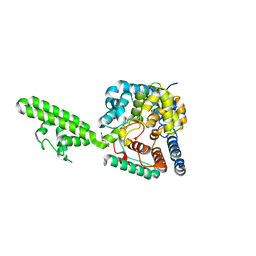 | | Structure of the Crimean-Congo Haemorrhagic Fever Virus Nucleocapsid Protein | | Descriptor: | NUCLEOCAPSID, TRIETHYLENE GLYCOL | | Authors: | Carter, S.D, Walter, C.T, Surtees, R, Bergeron, E, Ariza, A, Albarino, C.G, Nichol, S.T, Hiscox, J.A, Edwards, T.A, Barr, J.N. | | Deposit date: | 2012-02-24 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, Function, and Evolution of the Crimean-Congo Hemorrhagic Fever Virus Nucleocapsid Protein.
J.Virol., 86, 2012
|
|
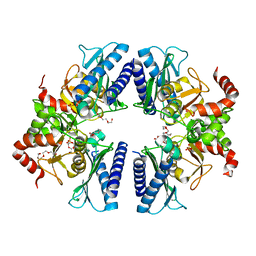
3T0J
 
 | |
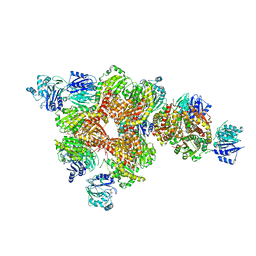
3OOJ
 
 | |
3GZ4
 
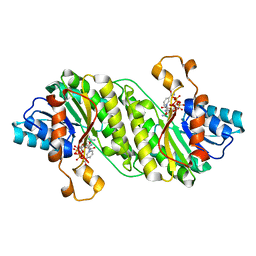 | | Crystal structure of putative short chain dehydrogenase FROM ESCHERICHIA COLI CFT073 complexed with NADPH | | Descriptor: | Hypothetical oxidoreductase yciK, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-06 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative short chain dehydrogenase
FROM ESCHERICHIA COLI CFT073 complexed with NADPH
To be Published
|
|
5WMK
 
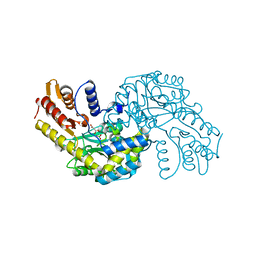 | |
3H1H
 
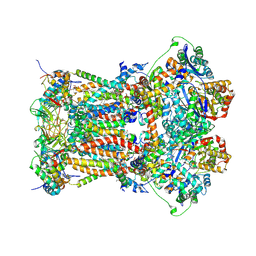 | | Cytochrome bc1 complex from chicken | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, CYTOCHROME C1, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 2009-04-12 | | Release date: | 2009-04-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
4ETW
 
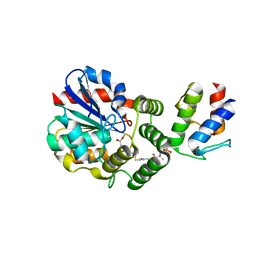 | | Structure of the Enzyme-ACP Substrate Gatekeeper Complex Required for Biotin Synthesis | | Descriptor: | Acyl carrier protein, Pimelyl-[acyl-carrier protein] methyl ester esterase, methyl 7-{[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-7-oxoheptanoate | | Authors: | Agarwal, V, Nair, S.K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-10-17 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the enzyme-acyl carrier protein (ACP) substrate gatekeeper complex required for biotin synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EY8
 
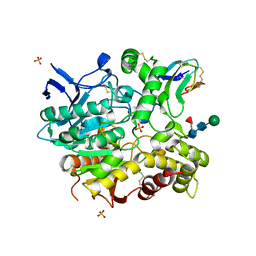 | | Crystal structure of recombinant human acetylcholinesterase in complex with fasciculin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, Fasciculin-2, ... | | Authors: | Cheung, J, Rudolph, M, Burshteyn, F, Cassidy, M, Gary, E, Love, J, Height, J, Franklin, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5958 Å) | | Cite: | Structures of human acetylcholinesterase in complex with pharmacologically important ligands.
J.Med.Chem., 55, 2012
|
|
4AGI
 
 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) in complex with seleno fucoside. | | Descriptor: | FUCOSE-SPECIFIC LECTIN FLEA, methyl 1-seleno-alpha-L-fucopyranoside | | Authors: | Houser, J, Komarek, J, Kostlanova, N, Lahmann, M, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2012-01-30 | | Release date: | 2013-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Soluble Fucose-Specific Lectin from Aspergillus Fumigatus Conidia - Structure, Specificity and Possible Role in Fungal Pathogenicity.
Plos One, 8, 2013
|
|
5X07
 
 | | Crystal structure of FOXA2 DNA binding domain bound to a full consensus DNA site | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Hepatocyte nuclear factor 3-beta | | Authors: | Li, J, Guo, M, Zhou, Z, Jiang, L, Chen, X, Qu, L, Wu, D, Chen, Z, Chen, L, Chen, Y. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site
Biochemistry, 56, 2017
|
|
5X1N
 
 | | Vanillate/3-O-methylgallate O-demethylase, LigM, protocatechuate-tetrahydrofolate complex form | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Harada, A, Senda, T. | | Deposit date: | 2017-01-26 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a new O-demethylase from Sphingobium sp. strain SYK-6
FEBS J., 284, 2017
|
|
