4EWJ
 
 | | structure of the enloase from Streptococcus suis serotype 2 | | Descriptor: | Enolase 2 | | Authors: | Lu, Q, Lu, H, Qi, J, Lu, G, Gao, G.F. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | An octamer of enolase from Streptococcus suis.
Protein Cell, 3, 2012
|
|
4E7B
 
 | |
4E8O
 
 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ih from Acinetobacter baumannii | | Descriptor: | Aac(6')-Ih protein, CHLORIDE ION | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
5J11
 
 | | Structure of human TSLP in complex with TSLPR and IL-7Ralpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor-like factor 2, Interleukin-7 receptor subunit alpha, ... | | Authors: | Verstraete, K, Savvides, S.N. | | Deposit date: | 2016-03-28 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure and antagonism of the receptor complex mediated by human TSLP in allergy and asthma.
Nat Commun, 8, 2017
|
|
1VYW
 
 | | Structure of CDK2/Cyclin A with PNU-292137 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, N-(3-CYCLOPROPYL-1H-PYRAZOL-5-YL)-2-(2-NAPHTHYL)ACETAMIDE, ... | | Authors: | Pevarello, P, Brasca, M.G, Amici, R, Orsini, P, Traquandi, G, Corti, L, Piutti, C, Sansonna, P, Villa, M, Pierce, B.S, Pulici, M, Giordano, P, Martina, K, Fritzen, E.L, Nugent, R.A, Casale, E, Cameron, A, Ciomei, M, Roletto, F, Isacchi, A, Fogliatto, G, Pesenti, E, Pastori, W, Marsiglio, A, Leach, K.L, Clare, P.M, Fiorentini, F, Varasi, M, Vulpetti, A, Warpehoski, M.A. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2/Cyclin a as Antitumor Agents. Part 1. Lead Finding
J.Med.Chem., 47, 2004
|
|
2NWZ
 
 | |
4ENY
 
 | | Crystal Structure of Pim-1 kinase in complex with (2E,5Z)-2-(2-chlorophenylimino)-5-(4-hydroxy-3-methoxybenzylidene)thiazolidin-4-one | | Descriptor: | (2Z,5Z)-2-[(2-chlorophenyl)imino]-5-(4-hydroxy-3-methoxybenzylidene)-1,3-thiazolidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2NXR
 
 | |
4DS1
 
 | |
5XUG
 
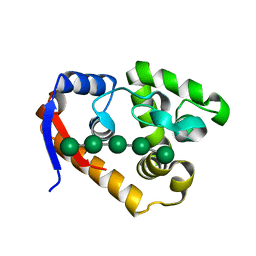 | | Complex structure(RmMan134A-M5). | | Descriptor: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, endo-1,4-beta-mannanase | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-06-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
1XRW
 
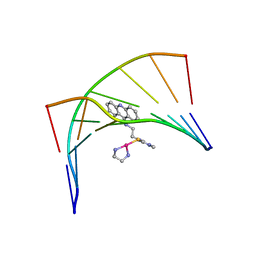 | | Solution Structure of a Platinum-Acridine Modified Octamer | | Descriptor: | 1-[2-(ACRIDIN-9-YLAMINO)ETHYL]-1,3-DIMETHYLTHIOUREA-PLATINUM(II)-ETHANE-1,2-DIAMINE, 5'-D(*CP*CP*TP*CP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*GP*AP*GP*G)-3' | | Authors: | Baruah, H, Wright, M.W, Bierbach, U. | | Deposit date: | 2004-10-16 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structural Study of a DNA Duplex Containing the Guanine-N7 Adduct Formed by a Cytotoxic Platinum-Acridine Hybrid Agent
Biochemistry, 44, 2005
|
|
2NWO
 
 | |
5JCA
 
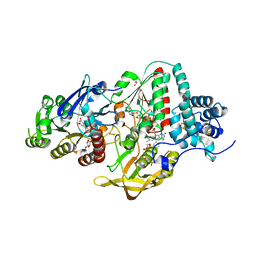 | | NADP(H) bound NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) from Pyrococcus furiosus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Zadvornyy, O.A, Schut, G.J, Nguyen, D.M, Artz, J.H, Tokmina-Lukaszewska, M, Lipscomb, G, Adams, M.W, Peters, J.W. | | Deposit date: | 2016-04-14 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic insights into energy conservation by flavin-based electron bifurcation.
Nat. Chem. Biol., 13, 2017
|
|
6G65
 
 | |
6G6C
 
 | |
5HNE
 
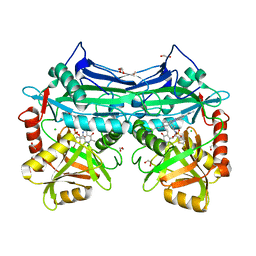 | | X-RAY CRYSTAL STRUCTURE OF HUMAN MITOCHONDRIAL BRANCHED CHAIN AMINOTRANSFERASE (BCATM) COMPLEXED WITH A 2-ARYL BENZIMIDAZOLE COMPOUND AND AN INTERNAL ALDIMINE LINKED PLP COFACTOR | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1R,3S)-3-{[(5-bromothiophen-2-yl)carbonyl]amino}cyclohexyl]-N-methyl-2-(pyridin-2-yl)-1H-benzimidazole-5-carboxamide, Branched-chain-amino-acid aminotransferase, ... | | Authors: | Somers, D.O. | | Deposit date: | 2016-01-18 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Optimization of Potent, Selective, and in Vivo Efficacious 2-Aryl Benzimidazole BCATm Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5XU5
 
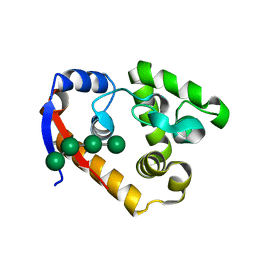 | | Complex structure of RmMan134A-M4 | | Descriptor: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, endo-1,4-beta-mannanase | | Authors: | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
7TZ3
 
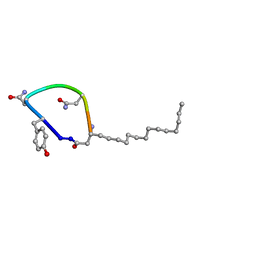 | |
6FTB
 
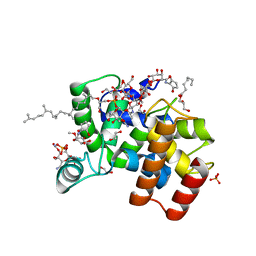 | | Staphylococcus aureus monofunctional glycosyltransferase in complex with moenomycin | | Descriptor: | (2R)-2,3-dihydroxypropyl dodecanoate, 1,2-ETHANEDIOL, MOENOMYCIN, ... | | Authors: | Punekar, A.S, Dowson, C.J, Roper, D.I. | | Deposit date: | 2018-02-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of the jaw subdomain of peptidoglycan glycosyltransferases for lipid II polymerization.
Cell Surf, 2, 2018
|
|
2NXS
 
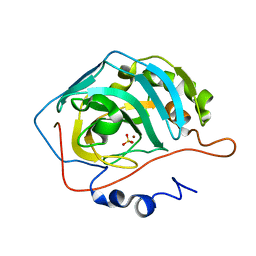 | |
1Y62
 
 | | A 2.4 crystal structure of conkunitzin-S1, a novel Kunitz-fold cone snail neurotoxin. | | Descriptor: | Conkunitzin-S1, SULFATE ION | | Authors: | Dy, C.Y, Buczek, P, Horvath, M.P. | | Deposit date: | 2004-12-03 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of conkunitzin-S1, a neurotoxin and Kunitz-fold disulfide variant from cone snail.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5Y7L
 
 | |
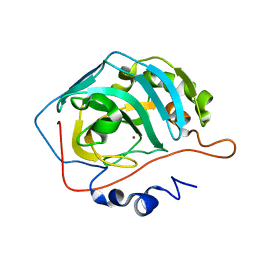
2NWY
 
 | |
4ERW
 
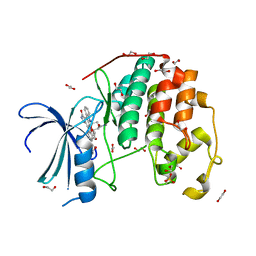 | | CDK2 in complex with staurosporine | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, STAUROSPORINE | | Authors: | Alam, R, Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
7OWZ
 
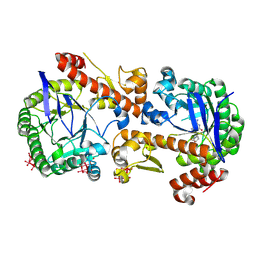 | | Heterodimeric murine tRNA-guanine transglycosylase in complex with queuine and in the presence of Anderson-Evans type (TEW) and Strandberg type polyoxometalate (POM) | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-tungstotellurate(VI), Queuine tRNA-ribosyltransferase accessory subunit 2, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
