7L7O
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR04-49 | | Descriptor: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
7LCA
 
 | |
7L7N
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-59 | | Descriptor: | 1,2-ETHANEDIOL, 1-(trifluoromethyl)cyclobutyl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3 protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
7L7L
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-129 | | Descriptor: | 1,1,1-trifluoro-2-methylpropan-2-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-{[6-methoxy-3-(trifluoromethyl)quinoxalin-2-yl]oxy}-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of Quinoxaline-Based P1-P3 Macrocyclic NS3/4A Protease Inhibitors with Potent Activity against Drug-Resistant Hepatitis C Virus Variants.
J.Med.Chem., 64, 2021
|
|
8DCC
 
 | |
7LB8
 
 | | Structure of a ferrichrome importer FhuCDB from E. coli | | Descriptor: | Iron(3+)-hydroxamate import ATP-binding protein FhuC, Iron(3+)-hydroxamate import system permease protein FhuB, Iron(3+)-hydroxamate-binding protein FhuD | | Authors: | Hu, W, Zheng, H. | | Deposit date: | 2021-01-07 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM reveals unique structural features of the FhuCDB Escherichia coli ferrichrome importer.
Commun Biol, 4, 2021
|
|
8T60
 
 | |
8D3F
 
 | | Crystal structure of human STAT1 in complex with the repeat region from Toxoplasma protein TgIST | | Descriptor: | Signal transducer and activator of transcription 1-alpha/beta,Inhibitor of STAT1-dependent transcription TgIST | | Authors: | Huang, Z, Liu, H, Nix, J.C, Amarasinghe, G.K, Sibley, L.D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | The intrinsically disordered protein TgIST from Toxoplasma gondii inhibits STAT1 signaling by blocking cofactor recruitment.
Nat Commun, 13, 2022
|
|
7LO9
 
 | | RNA dodecamer containing a GNA A residue | | Descriptor: | Chains: A,B,C,D | | Authors: | Harp, J.M, Wawrzak, Z, Egli, M. | | Deposit date: | 2021-02-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Overcoming GNA/RNA base-pairing limitations using isonucleotides improves the pharmacodynamic activity of ESC+ GalNAc-siRNAs.
Nucleic Acids Res., 49, 2021
|
|
7LUH
 
 | |
7LUJ
 
 | |
7LVZ
 
 | | Crystal structure of ADO | | Descriptor: | 2-aminoethanethiol dioxygenase, CHLORIDE ION, FE (II) ION, ... | | Authors: | Bingman, C.A, Fernandez, R.L, Smith, R.W, Fox, B.G, Brunold, T.C. | | Deposit date: | 2021-02-26 | | Release date: | 2022-01-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure of Cysteamine Dioxygenase Reveals the Origin of the Large Substrate Scope of This Vital Mammalian Enzyme.
Biochemistry, 60, 2021
|
|
7LCU
 
 | | X-ray structure of Furin bound to BOS-318, a small molecule inhibitor | | Descriptor: | (1-{[2-(3,5-dichlorophenyl)-6-{[2-(4-methylpiperazin-1-yl)pyrimidin-5-yl]oxy}pyridin-4-yl]methyl}piperidin-4-yl)acetic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Campobasso, N, Reid, R. | | Deposit date: | 2021-01-11 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A highly selective, cell-permeable furin inhibitor BOS-318 rescues key features of cystic fibrosis airway disease.
Cell Chem Biol, 29, 2022
|
|
7LTY
 
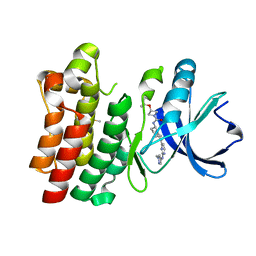 | | Bruton's tyrosine kinase in complex with compound 23 | | Descriptor: | DIMETHYL SULFOXIDE, Isoform BTK-C of Tyrosine-protein kinase BTK, ~{N}-[(5~{R})-2-[2-[(1-methylpyrazol-4-yl)amino]pyrimidin-4-yl]-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-5-yl]-3-propan-2-yloxy-azetidine-1-carboxamide | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2021-02-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB091, a Reversible, Selective BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 65, 2022
|
|
7LTZ
 
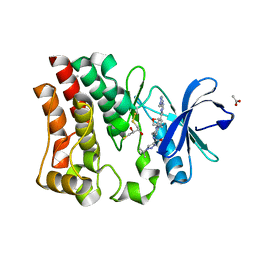 | | Bruton's tyrosine kinase in complex with compound 51 | | Descriptor: | 1-~{tert}-butyl-~{N}-[(5~{R})-8-[2-[(1-methylpyrazol-4-yl)amino]pyrimidin-4-yl]-2-(oxetan-3-yl)-1,3,4,5-tetrahydro-2-benzazepin-5-yl]-1,2,3-triazole-4-carboxamide, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, ... | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2021-02-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB091, a Reversible, Selective BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 65, 2022
|
|
7WBK
 
 | |
7LY5
 
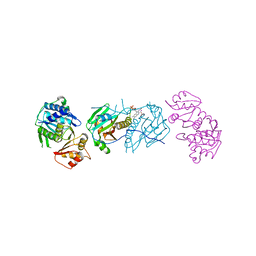 | | Proteolyzed crystal structure of the bacillamide NRPS, BmdB, in complex with the oxidase BmdC | | Descriptor: | BmdB, Bacillamide NRPS, BmdC, ... | | Authors: | Fortinez, C.M, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
7LY6
 
 | | Structure of a trans-acting NRPS oxidase, BmdC, involved in bacillamide biosynthesis | | Descriptor: | BmdC, NRPS oxidase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fortinez, C.M, Bloudoff, K, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
7U1Z
 
 | | Crystal structure of the DRBD and CROPs of TcdA | | Descriptor: | SULFATE ION, Toxin A | | Authors: | Baohua, C, Peng, C, Kay, P, Rongsheng, J. | | Deposit date: | 2022-02-22 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure and conformational dynamics of Clostridioides difficile toxin A.
Life Sci Alliance, 5, 2022
|
|
7LY7
 
 | | Crystal structure of the elongation module of the bacillamide NRPS, BmdB, in complex with the oxidase BmdC | | Descriptor: | 5'-{[(2R,3S)-3-amino-2-({2-[(N-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-beta-alanyl)amino]ethyl}sulfanyl)-4-sulfanylbutane-1-sulfonyl]amino}-5'-deoxyadenosine, BmdB, Bacillamide NRPS, ... | | Authors: | Fortinez, C.M, Sharon, I, Schmeing, T.M. | | Deposit date: | 2021-03-05 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module.
Nat Commun, 13, 2022
|
|
8D93
 
 | | [2T7] Self-assembling tensegrity triangle with R3 symmetry at 2.96 A resolution, update and junction cut for entry 3GBI | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*AP*CP*CP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Woloszyn, K, Lu, B, Sha, R, Ohayon, Y.P, Seeman, N.C. | | Deposit date: | 2022-06-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The Rule of Thirds: Controlling Junction Chirality and Polarity in 3D DNA Tiles.
Small, 19, 2023
|
|
8CXK
 
 | | Structure of the C. elegans HIM-3 R93Y mutant | | Descriptor: | HORMA domain-containing protein | | Authors: | Ego, K.M, Russo, A, Giacopazzi, S, Deshong, A, Menon, M, Ortiz, V, Bhalla, N, Corbett, K.D. | | Deposit date: | 2022-05-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The conserved AAA ATPase PCH-2 distributes its regulation of meiotic prophase events through multiple meiotic HORMADs in C. elegans.
Plos Genet., 19, 2023
|
|
7W9A
 
 | |
7LNH
 
 | | S-adenosylmethionine synthetase co-crystallized with UppNHp | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-$l^{3}-sulfanyl]butanoic acid, 1,2-ETHANEDIOL, S-adenosylmethionine synthase isoform type-2 | | Authors: | Tan, L.L, Jackson, C.J. | | Deposit date: | 2021-02-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
7M0E
 
 | | Pre-catalytic synaptic complex of DNA Polymerase Lambda with gapped DSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
