7T3U
 
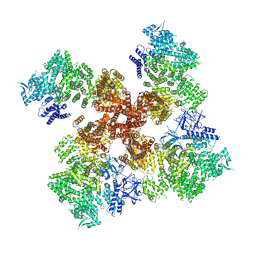 | | IP3, ATP, and Ca2+ bound type 3 IP3 receptor in the inactive state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3R
 
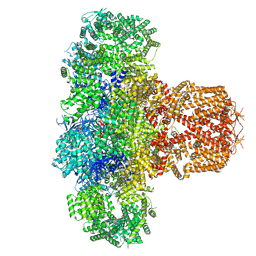 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active C state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
1K25
 
 | |
2VVM
 
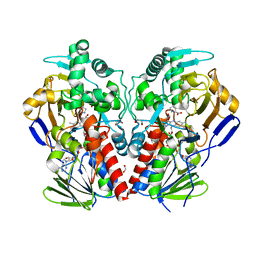 | | The structure of MAO-N-D5, a variant of monoamine oxidase from Aspergillus niger. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE N, ... | | Authors: | Atkin, K.E, Hart, S, Turkenburg, J.P, Brzozowski, A.M, Grogan, G.J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of Monoamine Oxidase from Aspergillus Niger Provides a Molecular Context for Improvements in Activity Obtained by Directed Evolution.
J.Mol.Biol., 384, 2008
|
|
1JG9
 
 | |
2XJF
 
 | |
1LKT
 
 | | CRYSTAL STRUCTURE OF THE HEAD-BINDING DOMAIN OF PHAGE P22 TAILSPIKE PROTEIN | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage.
J.Mol.Biol., 267, 1997
|
|
7T3P
 
 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active A state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
2VVL
 
 | | The structure of MAO-N-D3, a variant of monoamine oxidase from Aspergillus niger. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE N | | Authors: | Atkin, K.E, Hart, S, Turkenburg, J.P, Brzozowski, A.M, Grogan, G.J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Structure of Monoamine Oxidase from Aspergillus Niger Provides a Molecular Context for Improvements in Activity Obtained by Directed Evolution.
J.Mol.Biol., 384, 2008
|
|
4DA5
 
 | | Choline Kinase alpha acts through a double-displacement kinetic mechanism involving enzyme isomerisation, as determined through enzyme and inhibitor kinetics and structural biology | | Descriptor: | (3R)-1-azabicyclo[2.2.2]oct-3-yl[bis(5-chlorothiophen-2-yl)]methanol, Choline kinase alpha, SULFATE ION | | Authors: | Brown, K, Hudson, C, Charlton, P, Pollard, J. | | Deposit date: | 2012-01-12 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and mechanistic characterisation of Choline Kinase-alpha.
Biochim.Biophys.Acta, 1834, 2013
|
|
2XCW
 
 | |
1KV4
 
 | | Solution structure of antibacterial peptide (Moricin) | | Descriptor: | moricin | | Authors: | Hemmi, H, Ishibashi, J, Hara, S, Yamakawa, M. | | Deposit date: | 2002-01-25 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of moricin, an antibacterial peptide, isolated from the silkworm Bombyx mori.
FEBS Lett., 518, 2002
|
|
2XJB
 
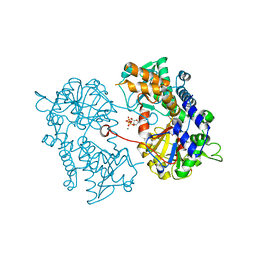 | |
2XCV
 
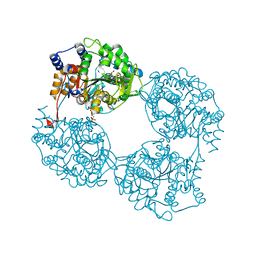 | | Crystal structure of the D52N variant of cytosolic 5'-nucleotidase II in complex with inosine monophosphate and 2,3-bisphosphoglycerate | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, CYTOSOLIC PURINE 5'-NUCLEOTIDASE, GLYCEROL, ... | | Authors: | Wallden, K, Nordlund, P. | | Deposit date: | 2010-04-26 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Allosteric Regulation and Substrate Recognition of Human Cytosolic 5'-Nucleotidase II
J.Mol.Biol., 408, 2011
|
|
2XJC
 
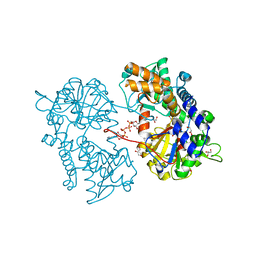 | |
2XJE
 
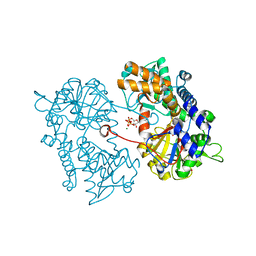 | |
4D3R
 
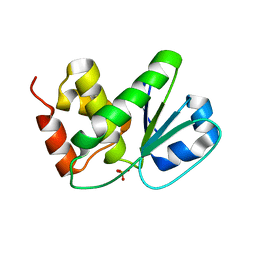 | |
4D3P
 
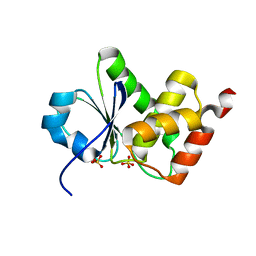 | |
4D3Q
 
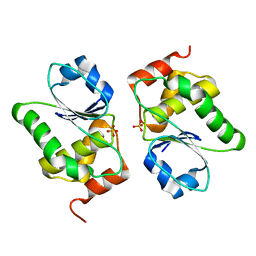 | |
2XCX
 
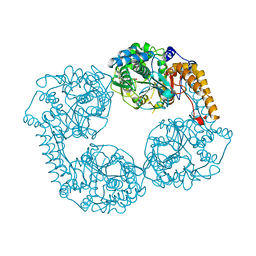 | |
2XJD
 
 | |
1JGI
 
 | |
1HKN
 
 | | A complex between acidic fibroblast growth factor and 5-amino-2-naphthalenesulfonate | | Descriptor: | 5-AMINO-NAPHTALENE-2-MONOSULFONATE, HEPARIN-BINDING GROWTH FACTOR 1 | | Authors: | Fernandez-Tornero, C, Lozano, R.M, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Leads for Development of New Naphthalenesulfonate Derivatives with Enhanced Antiangiogenic Activity: Crystal Structure of Acidic Fibroblast Growth Factor in Complex with 5-Amino-2-Naphthalenesulfonate
J.Biol.Chem., 278, 2003
|
|
1MO7
 
 | | ATPase | | Descriptor: | Sodium/Potassium-transporting ATPase alpha-1 chain | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
1MO8
 
 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Sodium/Potassium-Transporting ATPase alpha-1 | | Authors: | Hilge, M, Siegal, G, Vuister, G.W, Guentert, P, Gloor, S.M, Abrahams, J.P. | | Deposit date: | 2002-09-08 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ATP-induced conformational changes of the nucleotide-binding domain of Na,K-ATPase
Nat.Struct.Biol., 10, 2003
|
|
