1IE3
 
 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
2WKX
 
 | | Crystal structure of the native E. coli zinc amidase AmiD | | Descriptor: | CHLORIDE ION, GLYCEROL, N-ACETYLMURAMOYL-L-ALANINE AMIDASE AMID, ... | | Authors: | Petrella, S, Kerff, F, Herman, R, Genereux, C, Pennartz, A, Sauvage, E, Joris, B, Charlier, P. | | Deposit date: | 2009-06-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-L-Alanine Amidase Amid from Escherichia Coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
9D0X
 
 | | Cryo-EM structure of CDK2/CyclinE1 in complex with CRBN/DDB1 and Cpd 4 (local mask) | | Descriptor: | (3R)-3-(5-{4-[(2-{4-[(8-cyclopentyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl)amino]-3-methylbenzene-1-sulfonyl}-7-azaspiro[3.5]nonan-7-yl)methyl]piperidin-1-yl}-4-fluoro-3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1, ... | | Authors: | Kwiatkowski, N, Liang, T, Sha, Z, Collier, P.N, Yang, A, Sathappa, M, Paul, A, Su, L, Zheng, X, Aversa, R, Li, K, Mehovic, R, Breitkopf, S.B, Chen, D, Howarth, C.L, Yuan, K, Jo, H, Growney, J.D, Weiss, M, Williams, J. | | Deposit date: | 2024-08-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | CDK2 heterobifunctional degraders co-degrade CDK2 and cyclin E resulting in efficacy in CCNE1-amplified and overexpressed cancers.
Cell Chem Biol, 32, 2025
|
|
9D0W
 
 | | Cryo-EM structure of CDK2/CyclinE1 in complex with CRBN/DDB1 and Cpd 4 | | Descriptor: | (3R)-3-(5-{4-[(2-{4-[(8-cyclopentyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl)amino]-3-methylbenzene-1-sulfonyl}-7-azaspiro[3.5]nonan-7-yl)methyl]piperidin-1-yl}-4-fluoro-3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, Cyclin-dependent kinase 2, DNA damage-binding protein 1, ... | | Authors: | Kwiatkowski, N, Liang, T, Sha, Z, Collier, P.N, Yang, A, Sathappa, M, Paul, A, Su, L, Zheng, X, Aversa, R, Li, K, Mehovic, R, Breitkopf, S.B, Chen, D, Howarth, C.L, Yuan, K, Jo, H, Growney, J.D, Weiss, M, Williams, J. | | Deposit date: | 2024-08-07 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | CDK2 heterobifunctional degraders co-degrade CDK2 and cyclin E resulting in efficacy in CCNE1-amplified and overexpressed cancers.
Cell Chem Biol, 32, 2025
|
|
8V81
 
 | | Phosphorylated, ATP-bound, inhibitor 172-bound E1371Q human cystic fibrosis transmembrane conductance regulator | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(Z)-{(3M)-4-oxo-2-sulfanylidene-3-[3-(trifluoromethyl)phenyl]-1,3-thiazolidin-5-ylidene}methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gao, X, Hwang, T. | | Deposit date: | 2023-12-04 | | Release date: | 2024-07-17 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric inhibition of CFTR gating by CFTRinh-172 binding in the pore.
Nat Commun, 15, 2024
|
|
9GYA
 
 | | Vitamin D Receptor in complex with Sila-e | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R})-5-dimethylsilylpent-4-yn-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2024-10-01 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | First Sila-Vitamin D Analogues: Design, Synthesis, Structural Analysis and Biological Activity.
J.Med.Chem., 67, 2024
|
|
9C18
 
 | |
5L7I
 
 | | Structure of human Smoothened in complex with Vismodegib | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, SODIUM ION, ... | | Authors: | Byrne, E.X.B, Sircar, R, Miller, P.S, Hedger, G, Luchetti, G, Nachtergaele, S, Tully, M.D, Mydock-McGrane, L, Covey, D.F, Rambo, R.F, Sansom, M.S.P, Newstead, S, Rohatgi, R, Siebold, C. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of Smoothened regulation by its extracellular domains.
Nature, 535, 2016
|
|
2R29
 
 | | Neutralization of dengue virus by a serotype cross-reactive antibody elucidated by cryoelectron microscopy and x-ray crystallography | | Descriptor: | Envelope protein E, Heavy chain of Fab 1A1D-2, Light chain of Fab 1A1D-2 | | Authors: | Lok, S.M, Kostyuchenko, V.K, Nybakken, G.E, Holdaway, H.A, Battisti, A.J, Sukupolvi-petty, S, Sedlak, D, Fremont, D.H, Chipman, P.R, Roehrig, J.T, Diamond, M.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-08-24 | | Release date: | 2007-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
7Y8Y
 
 | | Structure of Cas7-11-crRNA-tgRNA in complex with TPR-CHAT | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (37-MER), ... | | Authors: | Wang, S, Guo, M, Zhu, Y, Huang, Z. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the type III-E CRISPR-Cas effector gRAMP in complex with TPR-CHAT.
Cell Res., 32, 2022
|
|
5LBR
 
 | | Wild-type PAS-GAF fragment from Deinococcus radiodurans Bphp collected at SACLA | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, Bacteriophytochrome | | Authors: | Edlund, P, Claesson, E, Nakane, T, Takala, H, Dods, R, Schmidt, M, Westenhoff, S. | | Deposit date: | 2016-06-17 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography.
Sci Rep, 6, 2016
|
|
6Y3B
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2110 | | Descriptor: | 1-[(8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, GLYCEROL, Genome polyprotein, ... | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
6NG0
 
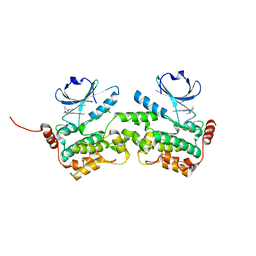 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with sunitinib in the inactive state. | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Johnson, E, McTigue, M, Cronin, C.N. | | Deposit date: | 2018-12-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Multiple conformational states of the HPK1 kinase domain in complex with sunitinib reveal the structural changes accompanying HPK1 trans-regulation.
J.Biol.Chem., 294, 2019
|
|
7ZOD
 
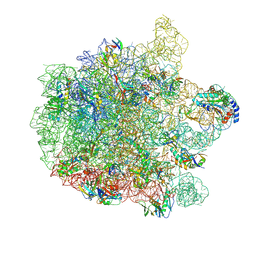 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
4RJY
 
 | |
1ZL6
 
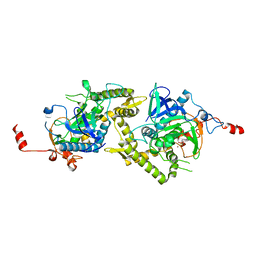 | | Crystal structure of Tyr350Ala mutant of Clostridium botulinum neurotoxin E catalytic domain | | Descriptor: | SULFATE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-05 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1EZF
 
 | | CRYSTAL STRUCTURE OF HUMAN SQUALENE SYNTHASE | | Descriptor: | FARNESYL-DIPHOSPHATE FARNESYLTRANSFERASE, N-{2-[TRANS-7-CHLORO-1-(2,2-DIMETHYL-PROPYL) -5-NAPHTHALEN-1-YL-2-OXO-1,2,3,5-TETRAHYDRO-BENZO[E] [1,4]OXAZEPIN-3-YL]-ACETYL}-ASPARTIC ACID | | Authors: | Pandit, J, Danley, D.E, Schulte, G.K, Mazzalupo, S.M, Pauly, T.A, Hayward, C.M, Hamanaka, E.S, Thompson, J.F, Harwood, H.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis.
J.Biol.Chem., 275, 2000
|
|
1F00
 
 | | CRYSTAL STRUCTURE OF C-TERMINAL 282-RESIDUE FRAGMENT OF ENTEROPATHOGENIC E. COLI INTIMIN | | Descriptor: | INTIMIN | | Authors: | Luo, Y, Frey, E.A, Pfuetzner, R.A, Creagh, A.L, Knoechel, D.G, Haynes, C.A, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2000-05-12 | | Release date: | 2000-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex.
Nature, 405, 2000
|
|
1NC3
 
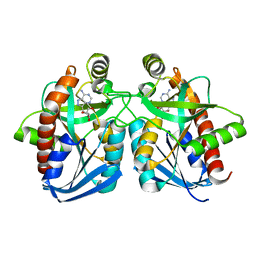 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with formycin A (FMA) | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2002-12-04 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
1CRZ
 
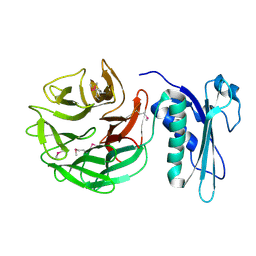 | | CRYSTAL STRUCTURE OF THE E. COLI TOLB PROTEIN | | Descriptor: | TOLB PROTEIN | | Authors: | Abergel, C, Bouveret, E, Claverie, J.-M, Brown, K, Rigal, A, Lazdunski, C, Benedetti, H. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution.
Structure Fold.Des., 7, 1999
|
|
8OV5
 
 | | PERIDININ-CHLOROPHYLL-PROTEIN OF AMPHIDINIUM CARTERAE, 100K | | Descriptor: | CHLOROPHYLL A, PERIDININ, Peridinin-chlorophyll a-binding protein 1, ... | | Authors: | Hofmann, E, Johanning, S. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and spectroscopic characterization of the peridinin-chlorophyll a-protein (PCP) complex from Heterocapsa pygmaea (HPPCP).
Biochim Biophys Acta Bioenerg, 1866, 2024
|
|
8OW6
 
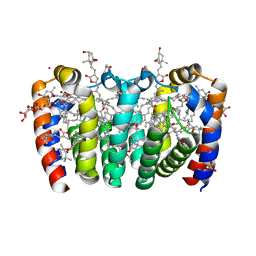 | | PERIDININ-CHLOROPHYLL-PROTEIN OF HETEROCAPSA PYGMAEA, 100K | | Descriptor: | CHLORIDE ION, CHLOROPHYLL A, PERIDININ, ... | | Authors: | Hofmann, E, Schulte, T. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and spectroscopic characterization of the peridinin-chlorophyll a-protein (PCP) complex from Heterocapsa pygmaea (HPPCP).
Biochim Biophys Acta Bioenerg, 1866, 2024
|
|
1GOS
 
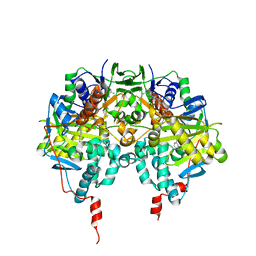 | | Human Monoamine Oxidase B | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE, N-[(E)-METHYL](PHENYL)-N-[(E)-2-PROPENYLIDENE]METHANAMINIUM | | Authors: | Binda, C, Newton-Vinson, P, Hubalek, F, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2001-10-26 | | Release date: | 2001-11-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Human Monoamine Oxidase B, a Drug Target for the Treatment of Neurological Disorders
Nat.Struct.Biol., 9, 2001
|
|
6D3Q
 
 | | Crystal structure of Escherichia coli enolase complexed with a natural inhibitor SF2312. | | Descriptor: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Erlandsen, H, Krucinska, J, Hazeen, A, Wright, D. | | Deposit date: | 2018-04-16 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
8XQN
 
 | | Structure of human class T GPCR TAS2R14-DNGi complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
