3DEC
 
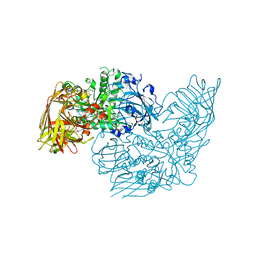 | | Crystal structure of a glycosyl hydrolases family 2 protein from Bacteroides thetaiotaomicron | | Descriptor: | Beta-galactosidase, POTASSIUM ION | | Authors: | Kumaran, D, Bonanno, J, Romero, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-09 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Glycosyl Hydrolases Family 2 protein from Bacteroides thetaiotaomicron.
To be Published
|
|
3VDC
 
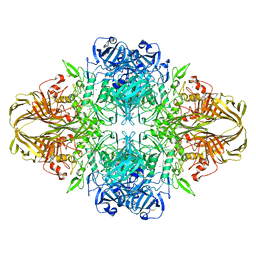 | | E. coli (lacZ) beta-galactosidase (N460T) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
3VD3
 
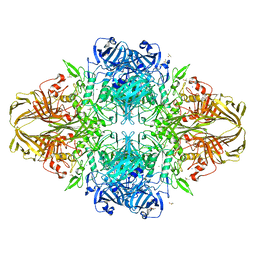 | | E. coli (lacZ) beta-galactosidase (N460D) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
3VD7
 
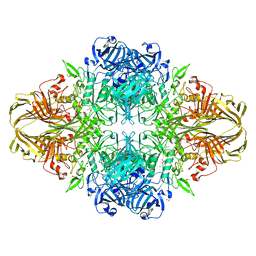 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with galactotetrazole | | Descriptor: | (5R, 6S, 7S, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
6LD0
 
 | | Structure of Bifidobacterium dentium beta-glucuronidase complexed with C6-hexyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-hexyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, LacZ1 Beta-galactosidase | | Authors: | Lin, H.-Y, Hsieh, T.-J, Lin, C.-H. | | Deposit date: | 2019-11-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LD6
 
 | |
6LDB
 
 | | Structure of Bifidobacterium dentium beta-glucuronidase complexed with uronic isofagomine | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, LacZ1 Beta-galactosidase | | Authors: | Lin, H.-Y, Hsieh, T.-J, Lin, C.-H. | | Deposit date: | 2019-11-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6XXW
 
 | | Structure of beta-D-Glucuronidase for Dictyoglomus thermophilum. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucuronidase, ... | | Authors: | Lafite, P, Daniellou, R. | | Deposit date: | 2020-01-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Thioglycoligation of aromatic thiols using a natural glucuronide donor.
Org.Biomol.Chem., 18, 2020
|
|
6LEM
 
 | | Structure of E. coli beta-glucuronidase complex with C6-nonyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-nonyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LDD
 
 | | Structure of Bifidobacterium dentium beta-glucuronidase complexed with C6-propyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)-2-propyl-piperidine-3-carboxylic acid, LacZ1 Beta-galactosidase | | Authors: | Lin, H.-Y, Hsieh, T.-J, Lin, C.-H. | | Deposit date: | 2019-11-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LEG
 
 | | Structure of E. coli beta-glucuronidase complex with uronic isofagomine | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LEL
 
 | | Structure of E. coli beta-glucuronidase complex with C6-hexyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-hexyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LEJ
 
 | | Structure of E. coli beta-glucuronidase complex with C6-propyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)-2-propyl-piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Lin, H.-Y, Kuo, Y.-H, Lin, C.-H. | | Deposit date: | 2019-11-25 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.617 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
6LDC
 
 | | Structure of Bifidobacterium dentium beta-glucuronidase complexed with C6-nonyl uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-2-nonyl-4,5-bis(oxidanyl)piperidine-3-carboxylic acid, LacZ1 Beta-galactosidase | | Authors: | Lin, H.-Y, Hsieh, T.-J, Lin, C.-H. | | Deposit date: | 2019-11-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Entropy-driven binding of gut bacterial beta-glucuronidase inhibitors ameliorates irinotecan-induced toxicity.
Commun Biol, 4, 2021
|
|
3SEP
 
 | | E. coli (lacZ) beta-galactosidase (S796A) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-06-10 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T0A
 
 | | E. coli (LacZ) beta-galactosidase (S796T) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T0B
 
 | | E. coli (LacZ) beta-galactosidase (S796T) IPTG complex | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | er-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T2O
 
 | | E. coli (lacZ) beta-galactosidase (S796D) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T09
 
 | | E. coli (LacZ) beta-galactosidase (S796A) galactonolactone complex | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T2Q
 
 | | E. coli (lacZ) beta-galactosidase (S796D) in complex with galactonolactone | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T08
 
 | | E. coli (LacZ) beta-galactosidase (S796A) IPTG complex | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T0D
 
 | | E.coli (lacZ) beta-galactosidase (S796T) in complex with galactonolactone | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ser-796 of Beta-galactosidase (E. coli) plays a Key role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T2P
 
 | | E. coli (lacZ) beta-galactosidase (S796D) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
6JZ5
 
 | |
6JZ8
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
