8SZO
 
 | | Canavalia villosa lectin in complex with alpha-methyl-mannoside | | Descriptor: | CALCIUM ION, Canavalia villosa lectin, GLYCEROL, ... | | Authors: | Cavada, B.S, Lossio, C.F, Pinto-Junior, V.R, Osterne, V.J.S, Oliveira, M.V, Neco, A.H.B, Nascimento, K.S. | | Deposit date: | 2023-05-30 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lectin from Canavalia villosa seeds: A glucose/mannose-specific protein and a new tool for inflammation studies.
Int J Biol Macromol, 105, 2017
|
|
6Z91
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC, SILVER ION | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
8EQ8
 
 | |
6Z99
 
 | | Copper transporter OprC | | Descriptor: | Putative copper transport outer membrane porin OprC, SILVER ION | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
6Z9Y
 
 | | Copper transporter OprC | | Descriptor: | COPPER (II) ION, Putative copper transport outer membrane porin OprC | | Authors: | Bhamidimarri, S.P, van den Berg, B. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site.
Plos Biol., 19, 2021
|
|
8EQH
 
 | |
6U54
 
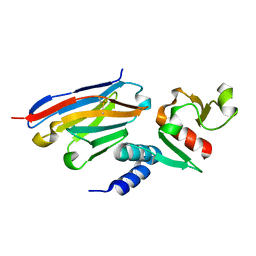 | | Anti-Zaire ebolavirus Nucleoprotein Single Domain Antibody Zaire C (ZC) Complexed with Zaire ebolavirus Nucleoprotein C-terminal Domain 634-739 | | Descriptor: | Anti-Zaire ebolavirus Nucleoprotein Single Domain Antibody Zaire C (ZC), Nucleoprotein | | Authors: | Taylor, A.B, Sherwood, L.J, Hart, P.J, Hayhurst, A. | | Deposit date: | 2019-08-26 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Paratope Duality and Gullying are Among the Atypical Recognition Mechanisms Used by a Trio of Nanobodies to Differentiate Ebolavirus Nucleoproteins.
J.Mol.Biol., 431, 2019
|
|
4TUP
 
 | |
8SO0
 
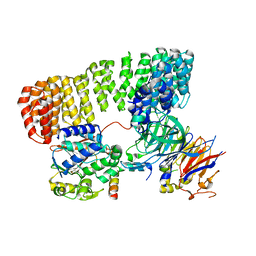 | | Cryo-EM structure of the PP2A:B55-FAM122A complex | | Descriptor: | FE (III) ION, PPP2R1A-PPP2R2A-interacting phosphatase regulator 1, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, ... | | Authors: | Fuller, J.R, Padi, S.K.R, Peti, W, Page, R. | | Deposit date: | 2023-04-28 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.79961 Å) | | Cite: | Cryo-EM structures of PP2A:B55-FAM122A and PP2A:B55-ARPP19.
Nature, 625, 2024
|
|
7NB5
 
 | |
6U4Y
 
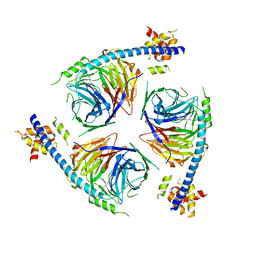 | | Crystal Structure of an EZH2-EED Complex in an Oligomeric State | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Jiao, L, Liu, X. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A partially disordered region connects gene repression and activation functions of EZH2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4U3Q
 
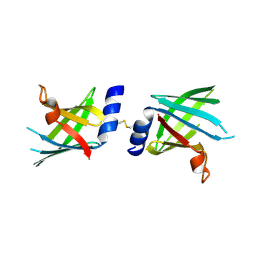 | |
8SMH
 
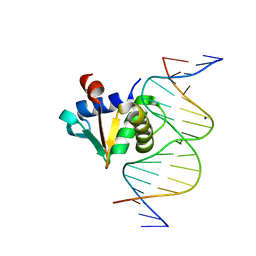 | |
8SP1
 
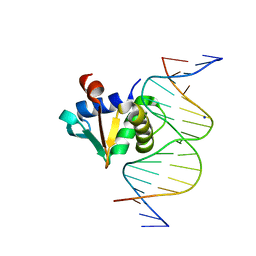 | |
8SMJ
 
 | |
8SJ5
 
 | | Walnut Tree Phytocystatin | | Descriptor: | Cysteine proteinase inhibitor | | Authors: | Valadares, N.F. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure and interconversion of monomers and domain-swapped dimers of the walnut tree phytocystatin.
Biochim Biophys Acta Proteins Proteom, 1872, 2023
|
|
4OTN
 
 | |
4UI1
 
 | | Crystal structure of the human RGMC-BMP2 complex | | Descriptor: | 1,2-ETHANEDIOL, BONE MORPHOGENETIC PROTEIN 2, CHLORIDE ION, ... | | Authors: | Healey, E.G, Bishop, B, Elegheert, J, Bell, C.H, Padilla-Parra, S, Siebold, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Repulsive Guidance Molecule is a Structural Bridge between Neogenin and Bone Morphogenetic Protein.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8SH3
 
 | | Pendrin in complex with iodide | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, IODIDE ION, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
8SGW
 
 | | Pendrin in complex with chloride | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
8SHC
 
 | | Pendrin in complex with Niflumic acid | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, CHLORIDE ION, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
8SIE
 
 | | Pendrin in complex with bicarbonate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, BICARBONATE ION, CHOLESTEROL, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-04-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
7N1O
 
 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Transcription factor ETV6,Isoform 4 of Anthrax toxin receptor 2 | | Authors: | Mathis, M.H, Bezzant, B.D, Ramirez, D.T, Sarath Nawarathnage, S.D, Doukov, T, Moody, J.D. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystals of TELSAM-target protein fusions that exhibit minimal crystal contacts and lack direct inter-TELSAM contacts.
Open Biology, 12, 2022
|
|
8SR6
 
 | |
4U7A
 
 | |
