4FZB
 
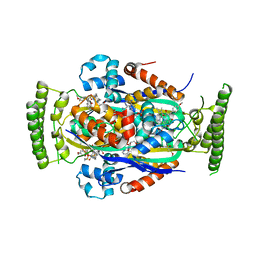 | | Structure of thymidylate synthase ThyX complexed to a new inhibitor | | Descriptor: | 2-hydroxy-3-(4-methoxybenzyl)naphthalene-1,4-dione, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Basta, T, Boum, Y, Briffotaux, J, Becker, H.F, Lamarre-Jouenne, I, Lambry, J.C, Skouloubris, S, Liebl, U, van Tilbeurgh, H, Graille, M, Myllylkallio, H. | | Deposit date: | 2012-07-06 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Mechanistic and structural basis for inhibition of thymidylate synthase ThyX.
Open Biology, 2, 2012
|
|
1UU8
 
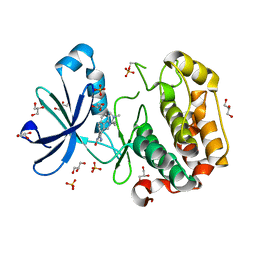 | | Structure of human PDK1 kinase domain in complex with BIM-1 | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Schuttelkopf, A.W, Deak, M, Prakash, K.R, Bain, J, Elliot, M, Garrido-Franco, M, Kozikowski, A.P, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-12-16 | | Release date: | 2004-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interactions of Ly333531 and Other Bisindolyl Maleimide Inhibitors with Pdk1
Structure, 12, 2004
|
|
1E95
 
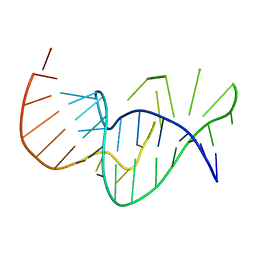 | | Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting | | Descriptor: | RNA (5'-(*GP*CP*GP*GP*CP*CP*AP*GP*CP*UP*CP* CP*AP*GP*GP*CP*CP*GP*CP*CP*AP*AP*AP*CP* AP*AP*UP*AP*UP*GP*GP*AP*GP*CP*AP*C)-3') | | Authors: | Michiels, P.J.A, Versleyen, A, Pleij, C.W.A, Hilbers, C.W, Heus, H.A. | | Deposit date: | 2000-10-09 | | Release date: | 2001-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pseudoknot of Srv-1 RNA, Involved in Ribosomal Frameshifting
J.Mol.Biol., 310, 2001
|
|
4K6O
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with 6-methyluracil at 1.17 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-methylpyrimidine-2,4-diol, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-04-16 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystallization and preliminary X-ray study of Vibrio cholerae uridine phosphorylase in complex with 6-methyluracil.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
5O9H
 
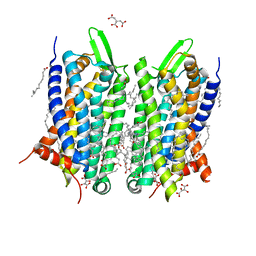 | | Crystal structure of thermostabilised human C5a anaphylatoxin chemotactic receptor 1 (C5aR) in complex with NDT9513727 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(1,3-benzodioxol-5-ylmethyl)-~{N}-[(3-butyl-2,5-diphenyl-imidazol-4-yl)methyl]methanamine, C5a anaphylatoxin chemotactic receptor 1, CITRIC ACID, ... | | Authors: | Robertson, N, Rappas, M, Dore, A.S, Brown, J, Bottegoni, G, Koglin, M, Cansfield, J, Jazayeri, A, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2017-06-19 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727.
Nature, 553, 2018
|
|
3EVX
 
 | | Crystal structure of the human E2-like ubiquitin-fold modifier conjugating enzyme 1 (Ufc1). Northeast Structural Genomics Consortium target HR41 | | Descriptor: | THIOCYANATE ION, Ufm1-conjugating enzyme 1 | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
4MX9
 
 | | CDPK1 from Neospora caninum in complex with inhibitor UW1294 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-[(1-methylpiperidin-4-yl)methyl]-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-like domain protein kinase isoenzyme gamma, related | | Authors: | Merritt, E.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
1CLK
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES DIASTATICUS NO.7 STRAIN M1033 XYLOSE ISOMERASE AT 1.9 A RESOLUTION WITH PSEUDO-I222 SPACE GROUP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Niu, L, Teng, M, Zhu, X, Gong, W. | | Deposit date: | 1999-04-29 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of xylose isomerase from Streptomyces diastaticus no. 7 strain M1033 at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2VGF
 
 | | HUMAN ERYTHROCYTE PYRUVATE KINASE: T384M mutant | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L.R, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-12 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
5NYM
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in reduced state | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SULFATE ION, ... | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
1OWO
 
 | | DATA4:photoreduced DNA photolyase / received X-rays dose 1.2 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2VGB
 
 | | HUMAN ERYTHROCYTE PYRUVATE KINASE | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-12 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
3UXL
 
 | |
5NYO
 
 | | Crystal structure of an atypical poplar thioredoxin-like2.1 variant in dimeric form | | Descriptor: | SULFATE ION, Thioredoxin-like protein 2.1 | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
1H10
 
 | | HIGH RESOLUTION STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN OF PROTEIN KINASE B/AKT BOUND TO INS(1,3,4,5)-TETRAKISPHOPHATE | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, RAC-ALPHA SERINE/THREONINE KINASE | | Authors: | Thomas, C.C, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2003-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High Resolution Structure of the Pleckstrin Homology Domain of Protein Kinase B/Akt Bound to Phosphatidylinositol (3,4,5)-Trisphosphate
Curr.Biol., 12, 2002
|
|
4AQP
 
 | | The structure of the AXH domain of ataxin-1. | | Descriptor: | ATAXIN-1, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Rees, M, Chen, Y.W, de Chiara, C, Pastore, A. | | Deposit date: | 2012-04-19 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Self-Assembly and Conformational Heterogeneity of the Axh Domain of Ataxin-1: An Unusual Example of a Chameleon Fold
Biophys.J., 104, 2013
|
|
2IO6
 
 | | Wee1 kinase complexed with inhibitor PD330961 | | Descriptor: | 9-HYDROXY-6-(3-HYDROXYPROPYL)-4-(2-METHOXYPHENYL)PYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2006-10-10 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases.
Eur.J.Med.Chem., 43, 2008
|
|
3PE6
 
 | |
7CU5
 
 | | N-Glycosylation of PD-1 and glycosylation dependent binding of PD-1 specific monoclonal antibody camrelizumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Programmed cell death protein 1, ... | | Authors: | Liu, K.F, Tan, S.G, Jin, W.J, Guan, J.W, Wang, W.L, Sun, H, Qi, J.X, Yan, J.H, Chai, Y, Wang, Z.F, Chu, X.D, Gao, G.F. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | N-glycosylation of PD-1 promotes binding of camrelizumab.
Embo Rep., 21, 2020
|
|
1D7K
 
 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE DECARBOXYLASE AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN ORNITHINE DECARBOXYLASE | | Authors: | Almrud, J.J, Oliveira, M.A, Kern, A.D, Grishin, N.V, Phillips, M.A, Hackert, M.L. | | Deposit date: | 1999-10-18 | | Release date: | 2000-10-25 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human ornithine decarboxylase at 2.1 A resolution: structural insights to antizyme binding.
J.Mol.Biol., 295, 2000
|
|
4F0Z
 
 | | Crystal Structure of Calcineurin in Complex with the Calcineurin-Inhibiting Domain of the African Swine Fever Virus Protein A238L | | Descriptor: | Ankyrin repeat domain-containing protein A238L, CALCIUM ION, Calcineurin subunit B type 1, ... | | Authors: | Grigoriu, S, Peti, W, Page, R. | | Deposit date: | 2012-05-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular mechanism of substrate engagement and immunosuppressant inhibition of calcineurin.
Plos Biol., 11, 2013
|
|
5NYK
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in oxidized state | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
6L0C
 
 | |
1I5X
 
 | | HIV-1 GP41 CORE | | Descriptor: | SULFATE ION, TRANSMEMBRANE GLYCOPROTEIN (GP41) | | Authors: | Liu, J, Lu, M. | | Deposit date: | 2001-03-01 | | Release date: | 2002-09-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of interhelical interactions in the human
immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning
mutagenesis.
J.Virol., 75, 2001
|
|
3UQF
 
 | | c-SRC kinase domain in complex with BKI RM-1-89 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Merritt, E.A, Larson, E.T. | | Deposit date: | 2011-11-20 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Multiple Determinants for Selective Inhibition of Apicomplexan Calcium-Dependent Protein Kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
