7K86
 
 | |
7YFQ
 
 | | Cryo-EM structure of the EfPiwi (N959K)-piRNA-target ternary complex | | Descriptor: | MAGNESIUM ION, Piwi, RNA (5'-R(*UP*CP*CP*AP*UP*GP*UP*UP*GP*AP*UP*GP*GP*UP*AP*A)-3'), ... | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
3NET
 
 | |
4DJJ
 
 | | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Pimelic acid at 2.9 Angstrom resolution | | Descriptor: | PIMELIC ACID, Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Singh, N, Sinha, M, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with Pimelic acid at 2.9 Angstrom resolution
To be Published
|
|
3J5S
 
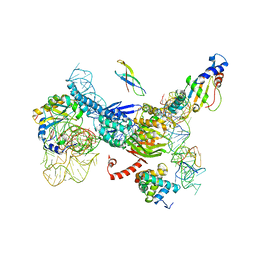 | |
1MMS
 
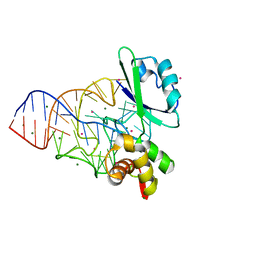 | | Crystal structure of the ribosomal PROTEIN L11-RNA complex | | Descriptor: | 23S RIBOSOMAL RNA, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Wimberly, B.T, Guymon, R, Mccutcheon, J.P, White, S.W, Ramakrishnan, V. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A detailed view of a ribosomal active site: the structure of the L11-RNA complex.
Cell(Cambridge,Mass.), 97, 1999
|
|
7ULZ
 
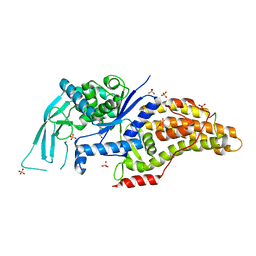 | |
4PQV
 
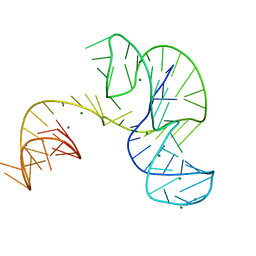 | | Crystal structure of an Xrn1-resistant RNA from the 3' untranslated region of a flavivirus (Murray Valley Encephalitis virus) | | Descriptor: | MAGNESIUM ION, XRN1-resistant flaviviral RNA | | Authors: | Chapman, E.G, Costantino, D.A, Rabe, J.L, Moon, S.L, Wilusz, J, Nix, J.C, Kieft, J.S. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production.
Science, 344, 2014
|
|
5AF0
 
 | | MAEL domain from Bombyx mori Maelstrom | | Descriptor: | MAELSTROM, ZINC ION | | Authors: | Chen, K, Campbell, E, Pandey, R.R, Yang, Z, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-01-13 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Metazoan Maelstrom is an RNA-Binding Protein that Has Evolved from an Ancient Nuclease Active in Protists.
RNA, 21, 2015
|
|
6URG
 
 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-CPSF100 PIM complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 4, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
5NOH
 
 | | HRSV M2-1 core domain | | Descriptor: | Transcription elongation factor M2-1 | | Authors: | Josts, I, Almeida Hernandez, Y, Molina, I.G, de Prat-Gay, G, Tidow, H. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the Human respiratory syncytial virus M2-1RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5NKX
 
 | | HRSV M2-1 core domain, P3221 crystal form | | Descriptor: | M2-1 | | Authors: | Almeida Hernandez, Y, Josts, I, Molina, I.G, de Pray-Gay, G, Tidow, H. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.00008678 Å) | | Cite: | Structure and stability of the Human respiratory syncytial virus M2-1RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4KR6
 
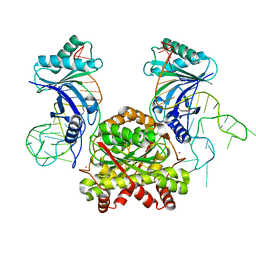 | |
4KR7
 
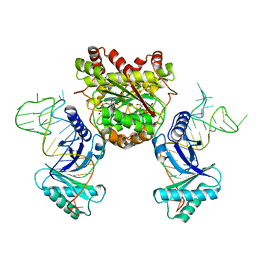 | | Crystal structure of a 4-thiouridine synthetase - RNA complex with bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable tRNA sulfurtransferase, ... | | Authors: | Neumann, P, Ficner, R, Lakomek, K. | | Deposit date: | 2013-05-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.421 Å) | | Cite: | Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification.
Nucleic Acids Res., 42, 2014
|
|
4KR9
 
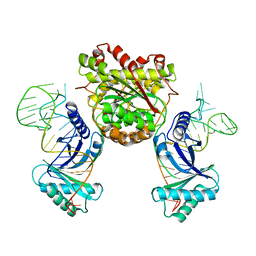 | |
5YYM
 
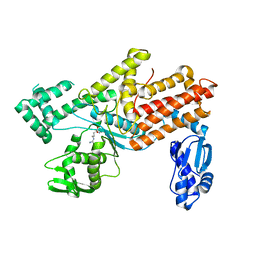 | | Crystal structures of E.coli arginyl-trna synthetase (argrs) in complex with substrate Arg | | Descriptor: | ARGININE, Arginine--tRNA ligase | | Authors: | Zhou, M, Ye, S, Stephen, P, Zhang, R.G, Wang, E.D, Giege, R, Lin, S.X. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures Of E.Coli Arginyl-Trna Synthetase (Argrs) In Complex With Substrates
To Be Published
|
|
3VBB
 
 | |
5BNZ
 
 | |
6FQ1
 
 | |
7YR8
 
 | | Lloviu cuevavirus nucleoprotein(1-450 residues)-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Hu, S.F, Fujita-Fujiharu, Y, Sugita, Y, Wendt, L, Muramoto, Y, Nakano, M, Hoenen, T, Noda, T. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryoelectron microscopic structure of the nucleoprotein-RNA complex of the European filovirus, Lloviu virus.
Pnas Nexus, 2, 2023
|
|
1FIR
 
 | |
7ODF
 
 | | Structure of the mini-RNA-guided endonuclease CRISPR-Cas_phi3 | | Descriptor: | Cas_phi3, DNA (5'-D(P*GP*G)-3'), DNA (5'-D(P*GP*TP*AP*AP*TP*TP*CP*AP*G)-3'), ... | | Authors: | Carabias del Rey, A, Fugilsang, A, Temperini, P, Pape, T, Sofos, N, Stella, S, Erledsson, S, Montoya, G. | | Deposit date: | 2021-04-29 | | Release date: | 2021-07-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the mini-RNA-guided endonuclease CRISPR-Cas12j3.
Nat Commun, 12, 2021
|
|
7YG6
 
 | | Cryo-EM structure of the EfPiwi(N959K) in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
7YGN
 
 | | Cryo-EM structure of the Mili in complex with piRNA | | Descriptor: | MAGNESIUM ION, Piwi-like protein 2, piRNA | | Authors: | Li, Z.Q, Liu, H.B, Wu, J.P, Shen, E.Z. | | Deposit date: | 2022-07-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing.
Nat.Struct.Mol.Biol., 2024
|
|
5IJX
 
 | |
