8PQG
 
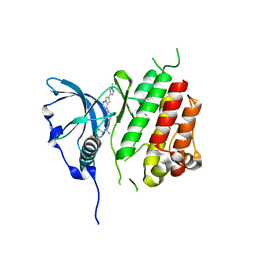 | | c-KIT T670I mutated kinase domain in complex with avapritinib | | Descriptor: | (1~{S})-1-(4-fluorophenyl)-1-[2-[4-[6-(1-methylpyrazol-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-yl]piperazin-1-yl]pyrimidin-5-yl]ethanamine, Mast/stem cell growth factor receptor Kit | | Authors: | Teuber, A, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-07-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Avapritinib-based SAR studies unveil a binding pocket in KIT and PDGFRA.
Nat Commun, 15, 2024
|
|
8PQD
 
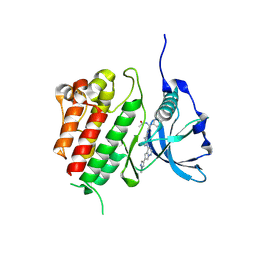 | | c-KIT kinase domain in complex with avapritinib derivative 10 | | Descriptor: | Mast/stem cell growth factor receptor Kit, ~{N}-[(1~{S})-1-(4-fluorophenyl)-1-[2-[4-[6-(1-methylpyrazol-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-yl]piperazin-1-yl]pyrimidin-5-yl]ethyl]ethanamide | | Authors: | Teuber, A, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-07-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Avapritinib-based SAR studies unveil a binding pocket in KIT and PDGFRA.
Nat Commun, 15, 2024
|
|
8PSX
 
 | |
8PT6
 
 | |
8PQK
 
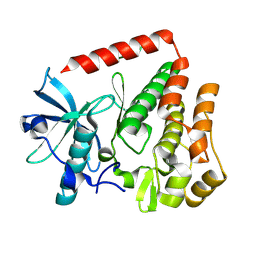 | |
8PSZ
 
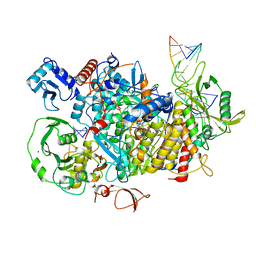 | |
6I56
 
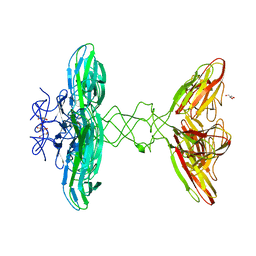 | | Crystal structure of PBSX exported protein XepA | | Descriptor: | GLYCEROL, Phage-like element PBSX protein XepA | | Authors: | Hakansson, M, Svensson, L.A, Welin, M, Al-Karadaghi, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structures of the Bacillus subtilis prophage lytic cassette proteins XepA and YomS.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RZH
 
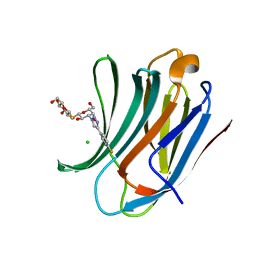 | | Galectin-3C in complex with para-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(4-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.947 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
6I6P
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 3 | | Descriptor: | 1,2-ETHANEDIOL, 6-azaspiro[3.4]octan-6-yl-[2,4-bis(chloranyl)-6-oxidanyl-phenyl]methanone, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
8Q7Y
 
 | |
8COP
 
 | |
8CQ8
 
 | |
8CQA
 
 | |
8CQ9
 
 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 5-(cyclopropylethynyl)-6-(4-(trifluoromethyl)phenyl)pyrimidine-2,4-diamine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(~{E})-2-cyclopropylethenyl]-6-[4-(trifluoromethyl)phenyl]pyrimidine-2,4-diamine, COBALT (II) ION, ... | | Authors: | Kirkman, T.J, Dias, M.V.B, Coyne, A.G. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Expansion of a series of pyrimidine derivatives utilising fragment-based merging leads to increased affinity to Mycobacterium tuberculosis dihydrofolate reductase (unpublished currently)
To Be Published
|
|
6I6C
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 2 | | Descriptor: | (1~{R},2~{S},4~{S})-~{N}-(3-chloranyl-4-cyano-phenyl)sulfonylbicyclo[2.2.1]heptane-2-carboxamide, 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | Descriptor: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-07-11 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6IBT
 
 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclophellitol aziridine ME737 | | Descriptor: | (1~{S},2~{S},3~{S},4~{S},5~{R},6~{S})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
6I5I
 
 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound 12h | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-[1-(phenylmethyl)pyrazol-4-yl]furo[3,2-b]pyridine, Dual specificity protein kinase CLK1 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6SDG
 
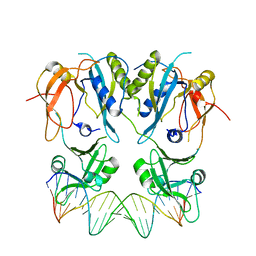 | |
8Q7E
 
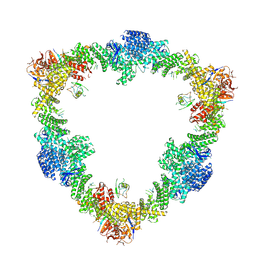 | |
6SFI
 
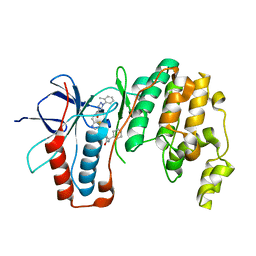 | | Crystal structure of p38 alpha in complex with compound 75 (MCP33) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]-2-[[[1-(2-methylphenyl)pyrazol-4-yl]carbonylamino]methyl]-1,3-thiazole-5-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
8Q7H
 
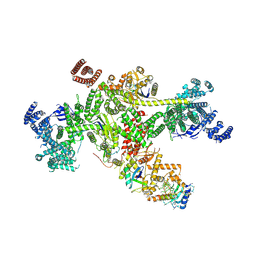 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, NEDD8, ... | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 2024
|
|
8CZI
 
 | |
8D27
 
 | |
6SFJ
 
 | | Crystal structure of p38 alpha in complex with compound 77 (MCP41) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-(2-methylphenyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
