2IWM
 
 | |
2IXV
 
 | |
6DI4
 
 | |
6O6C
 
 | | RNA polymerase II elongation complex arrested at a CPD lesion | | Descriptor: | DNA (27-MER), DNA (5'-D(P*GP*GP*AP*GP*AP*AP*GP*GP*AP*GP*CP*AP*GP*AP*GP*C)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Lahiri, I, Leshziner, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | 3.1 angstrom structure of yeast RNA polymerase II elongation complex stalled at a cyclobutane pyrimidine dimer lesion solved using streptavidin affinity grids.
J.Struct.Biol., 207, 2019
|
|
6OD7
 
 | | Herpes simplex virus type 1 (HSV-1) pUL6 portal protein, dodecameric complex | | Descriptor: | Portal protein | | Authors: | Liu, Y.T, Jih, J, Dai, X.H, Bi, G.Q, Zhou, Z.H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome.
Nature, 570, 2019
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JNF
 
 | | The cryo-EM structure of the RAD51 filament bound to the nucleosome | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.91 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
3LNX
 
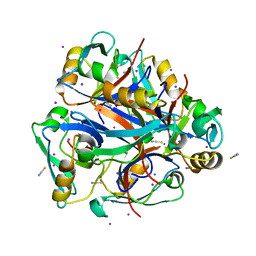 | | Second PDZ domain from human PTP1E | | Descriptor: | IODIDE ION, THIOCYANATE ION, Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Zhang, J, Chang, A, Ke, H, Phillips Jr, G.N, Lee, A.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-02-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystallographic and nuclear magnetic resonance evaluation of the impact of peptide binding to the second PDZ domain of protein tyrosine phosphatase 1E.
Biochemistry, 49, 2010
|
|
5XM0
 
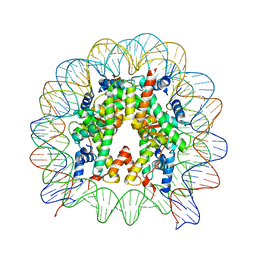 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3.3, and H4 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2017-05-12 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.874 Å) | | Cite: | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
5XNF
 
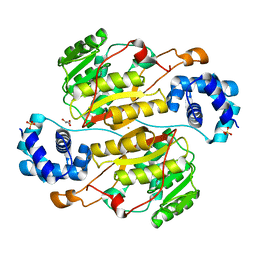 | | Crystal structure of the branched-chain polyamine synthase (BpsA) from Thermococcus kodakarensis | | Descriptor: | FE (III) ION, GLYCEROL, N(4)-bis(aminopropyl)spermidine synthase, ... | | Authors: | Mizohata, E, Tse, K.M, Fujita, J, Inoue, T. | | Deposit date: | 2017-05-22 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active site geometry of a novel aminopropyltransferase for biosynthesis of hyperthermophile-specific branched-chain polyamine.
FEBS J., 284, 2017
|
|
5K9M
 
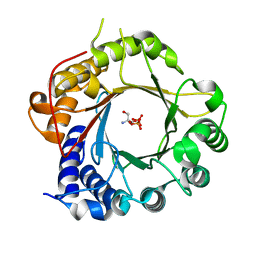 | | Crystal Structure of PriB Binary Complex with Product Diphosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYROPHOSPHATE 2-, PriB Prenyltransferase | | Authors: | Cao, H, Elshahawi, S, Benach, J, Wasserman, S.R, Morisco, L.L, Koss, J.W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2016-06-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and specificity of a permissive bacterial C-prenyltransferase.
Nat. Chem. Biol., 13, 2017
|
|
6OQQ
 
 | | Legionella pneumophila SidJ/Saccharomyces cerevisiae calmodulin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Black, M, Osinski, A. | | Deposit date: | 2019-04-28 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases.
Science, 364, 2019
|
|
4XUI
 
 | |
5XNH
 
 | | Crystal structure of the branched-chain polyamine synthase (BpsA) in complex with spermidine | | Descriptor: | FE (III) ION, N(4)-bis(aminopropyl)spermidine synthase, SPERMIDINE | | Authors: | Mizohata, E, Tse, K.M, Fujita, J, Inoue, T. | | Deposit date: | 2017-05-22 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Active site geometry of a novel aminopropyltransferase for biosynthesis of hyperthermophile-specific branched-chain polyamine.
FEBS J., 284, 2017
|
|
5J43
 
 | |
4KLB
 
 | | Crystal Structure of Cruzain in complex with the non-covalent inhibitor Nequimed176 | | Descriptor: | 2-{[(1H-1,2,4-triazol-5-ylsulfanyl)acetyl]amino}thiophene-3-carboxamide, Cruzipain | | Authors: | Fernandes, W.B, Montanari, C.A, Mckerrow, J.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Non-peptidic Cruzain Inhibitors with Trypanocidal Activity Discovered by Virtual Screening and In Vitro Assay.
Plos Negl Trop Dis, 7, 2013
|
|
6ESG
 
 | | Nucleosome breathing : Class 2 | | Descriptor: | DNA (141-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3LNY
 
 | | Second PDZ domain from human PTP1E in complex with RA-GEF2 peptide | | Descriptor: | Rap guanine nucleotide exchange factor 6, SULFATE ION, THIOCYANATE ION, ... | | Authors: | Zhang, J, Chang, A, Ke, H, Phillips Jr, G.N, Lee, A.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-02-03 | | Release date: | 2010-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic and nuclear magnetic resonance evaluation of the impact of peptide binding to the second PDZ domain of protein tyrosine phosphatase 1E.
Biochemistry, 49, 2010
|
|
6ESH
 
 | | Nucleosome breathing : Class 3 | | Descriptor: | DNA (137-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5XNC
 
 | | Crystal structure of the branched-chain polyamine synthase (BpsA) in complex with N4-aminopropylspermidine and 5-methylthioadenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mizohata, E, Tse, K.M, Fujita, J, Inoue, T. | | Deposit date: | 2017-05-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Active site geometry of a novel aminopropyltransferase for biosynthesis of hyperthermophile-specific branched-chain polyamine.
FEBS J., 284, 2017
|
|
6ESI
 
 | | Nucleosome breathing : Class 4 | | Descriptor: | DNA (133-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5XY3
 
 | | Large subunit of Trichomonas vaginalis ribosome | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Z, Guo, Q, Zheng, L, Ji, Y, Xie, Y, Lai, D, Lun, Z, Suo, X, Gao, N. | | Deposit date: | 2017-07-06 | | Release date: | 2017-08-30 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii
Cell Res., 27, 2017
|
|
8FKY
 
 | |
7KLP
 
 | | Crystal structure of a three-tetrad, parallel, K+ stabilized human telomeric G-quadruplex | | Descriptor: | HEXAETHYLENE GLYCOL, POTASSIUM ION, telomeric DNA | | Authors: | Yatsunyk, L.A, Li, K.S, Chen, E.V. | | Deposit date: | 2020-10-30 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Water spines and networks in G-quadruplex structures.
Nucleic Acids Res., 49, 2021
|
|
