3ZN1
 
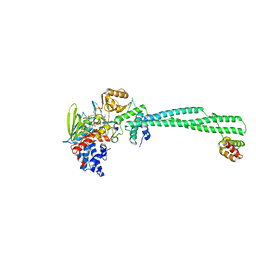 | | LSD1-CoREST in complex with PRLYLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZMV
 
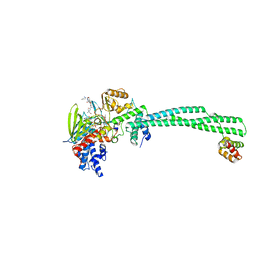 | | LSD1-CoREST in complex with PLSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PKSFLV PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3PXP
 
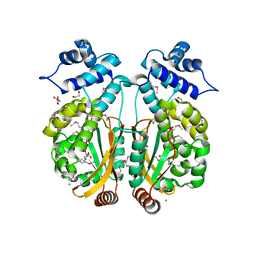 | |
2NZU
 
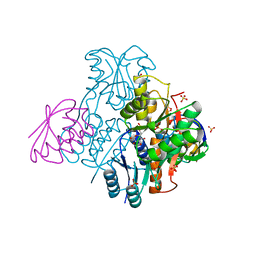 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | Authors: | Schumacher, M.A, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-11-25 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
6KGL
 
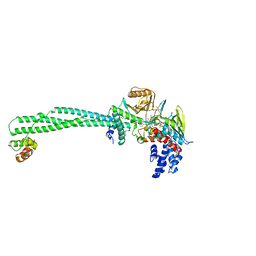 | | LSD1-CoREST-S2101 N5 adduct model | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Sengoku, S, Umehara, T, Yokoyama, S. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
6KGN
 
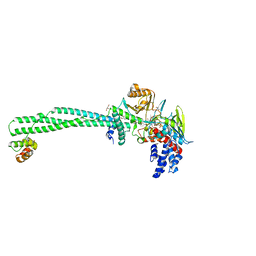 | | LSD1-CoREST-S2116 N5 adduct model | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
2P1N
 
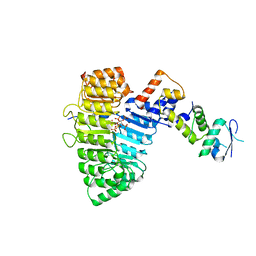 | | Mechanism of Auxin Perception by the TIR1 Ubiqutin Ligase | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, C, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
2P1O
 
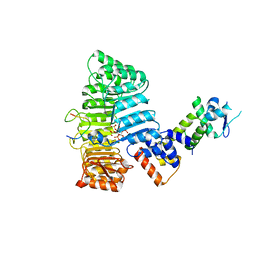 | | Mechanism of Auxin Perception by the TIR1 ubiquitin ligase | | Descriptor: | Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, NAPHTHALEN-1-YL-ACETIC ACID, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
3OHU
 
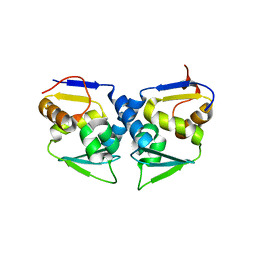 | | Crystal structure of the human Bach2 POZ domain, form I | | Descriptor: | Transcription regulator protein BACH2 | | Authors: | Rosbrook, G.O, Stead, M.A, Carr, S.B, Wright, S.C. | | Deposit date: | 2010-08-18 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the Bach2 POZ-domain dimer reveals an intersubunit disulfide bond.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3OHV
 
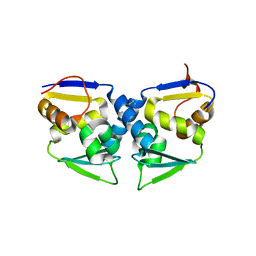 | | Crystal structure of the human Bach2 POZ domain, form II | | Descriptor: | Transcription regulator protein BACH2 | | Authors: | Rosbrook, G.O, Stead, M.A, Carr, S.B, Wright, S.C. | | Deposit date: | 2010-08-18 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the Bach2 POZ-domain dimer reveals an intersubunit disulfide bond.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1HNE
 
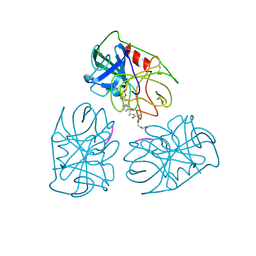 | | Structure of human neutrophil elastase in complex with a peptide chloromethyl ketone inhibitor at 1.84-angstroms resolution | | Descriptor: | HUMAN LEUCOCYTE ELASTASE, METHOXYSUCCINYL-ALA-ALA-PRO-ALA CHLOROMETHYL KETONE INHIBITOR | | Authors: | Navia, M.A, Mckeever, B.M, Springer, J.P, Lin, T.-Y, Williams, H.R, Fluder, E.M, Dorn, C.P, Hoogsteen, K. | | Deposit date: | 1989-04-10 | | Release date: | 1989-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of human neutrophil elastase in complex with a peptide chloromethyl ketone inhibitor at 1.84-A resolution.
Proc.Natl.Acad.Sci.USA, 86, 1989
|
|
2BKA
 
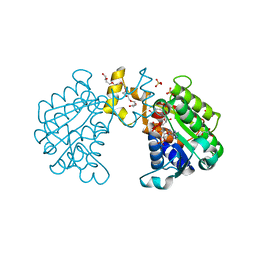 | | CC3(TIP30)Crystal Structure | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | El Omari, K, Bird, L.E, Nichols, C.E, Ren, J, Stammers, D.K. | | Deposit date: | 2005-02-14 | | Release date: | 2005-02-21 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Cc3 (Tip30): Implications for its Role as a Tumor Suppressor
J.Biol.Chem., 280, 2005
|
|
3HTH
 
 | |
3HTJ
 
 | |
3HTA
 
 | |
1E91
 
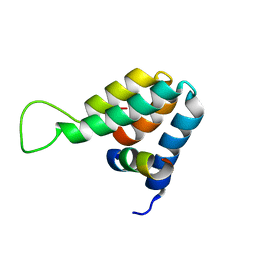 | | Structure of the complex of the Mad1-Sin3B interaction domains | | Descriptor: | MAD PROTEIN (MAX DIMERIZER), PAIRED AMPHIPATHIC HELIX PROTEIN SIN3B | | Authors: | Spronk, C.A.E.M, Tessari, M, Kaan, A.M, Jansen, J.F.A, Vermeulen, M, Stunnenberg, H.G, Vuister, G.W. | | Deposit date: | 2000-10-04 | | Release date: | 2000-11-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MAD1-Sin3B Interaction Involves a Novel Helical Fold
Nat.Struct.Biol., 7, 2000
|
|
1NKD
 
 | |
2NZV
 
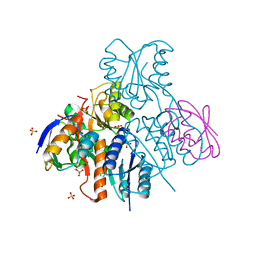 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | Authors: | Schumacher, M.A, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-11-25 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
5Y4T
 
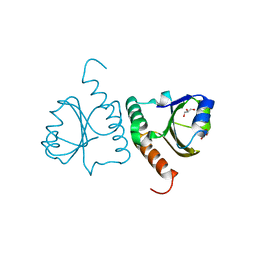 | | Crystal structure of Trx domain of Grx3 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Glutaredoxin | | Authors: | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-05 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
8CEO
 
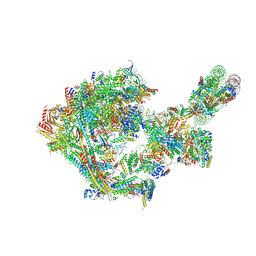 | |
8CEN
 
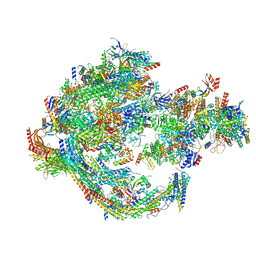 | | Yeast RNA polymerase II transcription pre-initiation complex with core Mediator | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, H, Schilbach, S, Cramer, P. | | Deposit date: | 2023-02-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Yeast PIC-Mediator structure with RNA polymerase II C-terminal domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5YDD
 
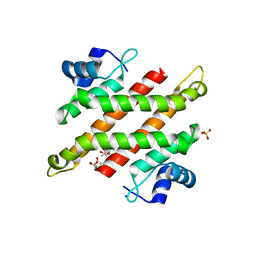 | | Crystal structure of C-terminal domain of Rv1828 from Mycobacterium tuberculosis | | Descriptor: | (6R,8S,9S)-8-(hydroxymethyl)-6,11,11-tris(oxidanyl)-9-propyl-dodecanoic acid, GLYCEROL, SODIUM ION, ... | | Authors: | Singh, S, Karthiekeyan, S. | | Deposit date: | 2017-09-12 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characteristics of the essential pathogenicity factor Rv1828, a MerR family transcription regulator from Mycobacterium tuberculosis.
FEBS J., 285, 2018
|
|
5Z4D
 
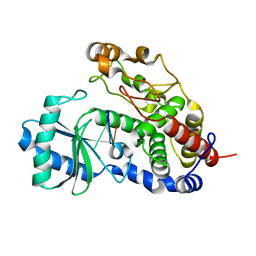 | | Structure of Tailor in complex with AGUU RNA | | Descriptor: | RNA (5'-R(*AP*GP*UP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4M
 
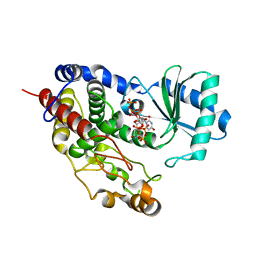 | | Structure of TailorD343A with bound UTP and Mg | | Descriptor: | MAGNESIUM ION, Terminal uridylyltransferase Tailor, URIDINE 5'-TRIPHOSPHATE | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
5Z4C
 
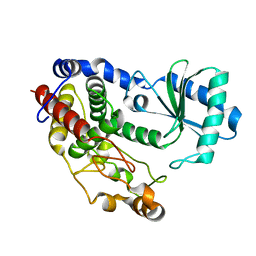 | | Crystal structure of Tailor | | Descriptor: | Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
