8UQM
 
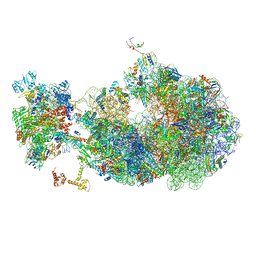 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH in loaded state, NusA, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-24 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8URI
 
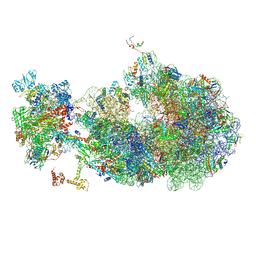 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, NusA, mRNA with a 27 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8URX
 
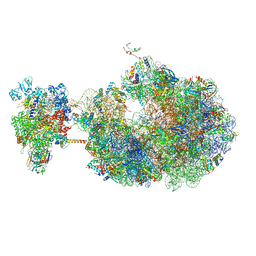 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, mRNA with a 30 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-27 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UR0
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, NusA, mRNA with a 24 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-25 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8UPR
 
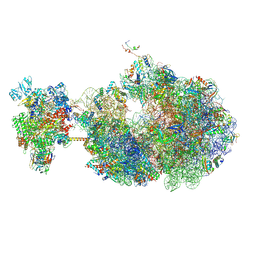 | | Escherichia coli transcription-translation coupled complex class A (TTC-A) containing RfaH bound to ops signal, mRNA with a 21 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
8URY
 
 | | Escherichia coli transcription-translation coupled complex class B (TTC-B) containing RfaH bound to ops signal, NusA, mRNA with a 30 nt long spacer, and fMet-tRNA in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
6YMW
 
 | |
8UPO
 
 | | Escherichia coli transcription-translation coupled complex class A (TTC-A) containing RfaH bound to ops signal, mRNA with a 21 nt long spacer, and fMet-tRNAs in E-site and P-site of the ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2023-10-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural basis of RfaH-mediated transcription-translation coupling.
Nat.Struct.Mol.Biol., 2024
|
|
7PLO
 
 | | H. sapiens replisome-CUL2/LRR1 complex | | Descriptor: | Cell division control protein 45 homolog, Claspin, Cullin-2, ... | | Authors: | Jones, M.J, Yeeles, J.T.P, Deegan, T.D, Jenkyn-Bedford, M. | | Deposit date: | 2021-09-01 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | Authors: | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | Deposit date: | 2009-08-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19.799999 Å) | | Cite: | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7EH0
 
 | |
7EH1
 
 | | Thermus thermophilus transcription initiation complex containing a template-strand purine at position TSS-2, GpG RNA primer, and CMPcPP | | Descriptor: | 1,4-BUTANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (27-MER), ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Promoter-sequence determinants and structural basis of primer-dependent transcription initiation in Escherichia coli .
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5OA1
 
 | | RNA polymerase I pre-initiation complex | | Descriptor: | ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Sadian, Y, Tafur, L, Kosinski, J, Jakobi, A.J, Muller, C.W. | | Deposit date: | 2017-06-20 | | Release date: | 2017-07-26 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into transcription initiation by yeast RNA polymerase I.
EMBO J., 36, 2017
|
|
6GOV
 
 | | Structure of THE RNA POLYMERASE LAMBDA-BASED ANTITERMINATION COMPLEX | | Descriptor: | 30S ribosomal protein S10, Antitermination protein N, DNA (I), ... | | Authors: | Loll, B, Krupp, F, Said, N, Huang, Y, Buerger, J, Mielke, T, Spahn, C.M.T, Wahl, M.C. | | Deposit date: | 2018-06-04 | | Release date: | 2019-02-13 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for the Action of an All-Purpose Transcription Anti-termination Factor.
Mol.Cell, 74, 2019
|
|
8W8P
 
 | |
4C3I
 
 | | Structure of 14-subunit RNA polymerase I at 3.0 A resolution, crystal form C2-100 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, ... | | Authors: | Fernandez-Tornero, C, Moreno-Morcillo, M, Rashid, U.J, Taylor, N.M.I, Ruiz, F.M, Gruene, T, Legrand, P, Steuerwald, U, Muller, C.W. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 14-Subunit RNA Polymerase I
Nature, 502, 2013
|
|
7VBA
 
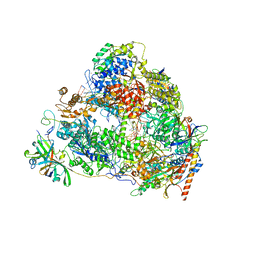 | | Structure of the pre state human RNA Polymerase I Elongation Complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (5'-D(P*A*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*AP*GP*AP*GP*AP*CP*AP*GP*CP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), ... | | Authors: | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
5DMA
 
 | |
8BYQ
 
 | | RNA polymerase II pre-initiation complex with the proximal +1 nucleosome (PIC-Nuc10W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
8XP8
 
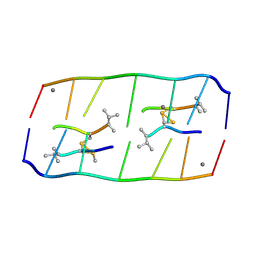 | | Crystal structure of d(ACGmCCGT/ACGGCGT) in complex with Echinomycin | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*(5CM)P*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3'), ... | | Authors: | Hou, M.H, Lin, S.M, Neidle, H. | | Deposit date: | 2024-01-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
1FIA
 
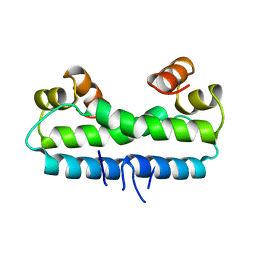 | | CRYSTAL STRUCTURE OF THE FACTOR FOR INVERSION STIMULATION FIS AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Kostrewa, D, Granzin, J, Choe, H.-W, Labahn, J, Saenger, W. | | Deposit date: | 1991-12-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the factor for inversion stimulation FIS at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
8C8H
 
 | |
8URW
 
 | | Cyanobacterial RNA polymerase elongation complex with NusG and CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA (40-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Qayyum, M.Z, Imashimizu, M, Leanca, M, Vishwakarma, R.K, Bradley Riaz, A, Yuzenkova, Y, Murakami, K.S. | | Deposit date: | 2023-10-26 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structure and function of the Si3 insertion integrated into the trigger loop/helix of cyanobacterial RNA polymerase.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2VUM
 
 | | Alpha-amanitin inhibited complete RNA polymerase II elongation complex | | Descriptor: | 5'-D(*AP*AP*AP*CP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *GP*TP*TP*AP*CP*GP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*AP*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Brueckner, F, Cramer, P. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Transcription Inhibition by Alpha-Amanitin and Implications for RNA Polymerase II Translocation.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8WAT
 
 | | De novo transcribing complex 10 (TC10), the early elongation complex with Pol II positioned 10nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
