6K0N
 
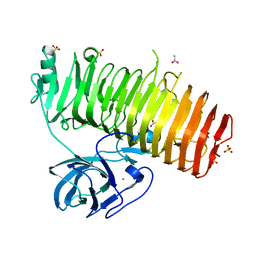 | | Catalytic domain of GH87 alpha-1,3-glucanase in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
4CCT
 
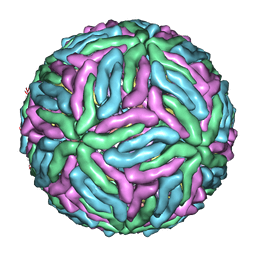 | | Dengue 1 cryo-EM reconstruction | | Descriptor: | DENGUE VIRUS 1 E PROTEIN, DENGUE VIRUS 1 M PROTEIN | | Authors: | Kostyuchenko, V.A, Zhang, Q, Tan, J.L, Ng, T.S, Lok, S.M. | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Immature and Mature Dengue Serotype 1 Virus Structures Provide Insight Into the Maturation Process.
J.Virol., 87, 2013
|
|
2R8O
 
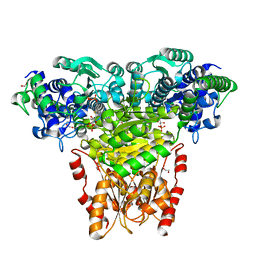 | | Transketolase from E. coli in complex with substrate D-xylulose-5-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-C-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thia zol-3-ium-2-yl}-5-O-phosphono-D-xylitol, CALCIUM ION, ... | | Authors: | Wille, G, Asztalos, P, Weiss, M.S, Tittmann, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Strain and near attack conformers in enzymic thiamin catalysis: X-ray crystallographic snapshots of bacterial transketolase in covalent complex with donor ketoses xylulose 5-phosphate and fructose 6-phosphate, and in noncovalent complex with acceptor aldose ribose 5-phosphate.
Biochemistry, 46, 2007
|
|
3UG3
 
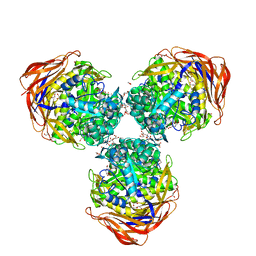 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, ... | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
2RK4
 
 | | Structure of M26I DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Lakshminarasimhan, M, Maldonado, M.T, Zhou, W, Fink, A.L, Wilson, M.A. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Impact of Three Parkinsonism-Associated Missense Mutations on Human DJ-1.
Biochemistry, 47, 2008
|
|
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | Descriptor: | Programmed cell death protein 1 | | Authors: | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
2MQ4
 
 | |
4OTQ
 
 | | Crystal structure of BTK kinase domain complexed with 1-[5-[3-(7-tert-butyl-4-oxo-quinazolin-3-yl)-2-methyl-phenyl]-1-methyl-2-oxo-3-pyridyl]-3-methyl-urea | | Descriptor: | 1-{5-[3-(7-tert-butyl-4-oxoquinazolin-3(4H)-yl)-2-methylphenyl]-1-methyl-2-oxo-1,2-dihydropyridin-3-yl}-3-methylurea, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Kuglstatter, A, Wong, A. | | Deposit date: | 2014-02-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Drug Design of RN486, a Potent and Selective Bruton's Tyrosine Kinase (BTK) Inhibitor, for the Treatment of Rheumatoid Arthritis.
J.Med.Chem., 58, 2015
|
|
1NU3
 
 | | Limonene-1,2-epoxide hydrolase in complex with valpromide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PROPYLPENTANAMIDE, limonene-1,2-epoxide hydrolase | | Authors: | Arand, M, Hallberg, B.M, Zou, J, Bergfors, T, Oesch, F, van der Werf, M.J, de Bont, J.A.M, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Rhodococcus erythropolis limonene-1,2-epoxide hydrolase reveals a novel active site
EMBO J., 22, 2003
|
|
4OLY
 
 | |
6K0S
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
4OR2
 
 | | Human class C G protein-coupled metabotropic glutamate receptor 1 in complex with a negative allosteric modulator | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-fluoro-N-methyl-N-{4-[6-(propan-2-ylamino)pyrimidin-4-yl]-1,3-thiazol-2-yl}benzamide, CHOLESTEROL, ... | | Authors: | Wu, H, Wang, C, Gregory, K.J, Han, G.W, Cho, H.P, Xia, Y, Niswender, C.M, Katritch, V, Cherezov, V, Conn, P.J, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator
Science, 344, 2014
|
|
3LAS
 
 | | Crystal structure of carbonic anhydrase from streptococcus mutans to 1.4 angstrom resolution | | Descriptor: | GLYCEROL, GUANIDINE, MAGNESIUM ION, ... | | Authors: | Ma, L.-L, Wang, K.-T, Liu, X, Su, X.-D. | | Deposit date: | 2010-01-07 | | Release date: | 2011-01-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of carbonic anhydrase from streptococcus mutans to 1.4 angstrom resolution
To be published
|
|
2MSN
 
 | | NMR structure of a putative phosphoglycolate phosphatase (NP_346487.1) from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
3W0W
 
 | | The complex between T36-5 TCR and HLA-A24 bound to HIV-1 Nef134-10(2F) peptide in space group P212121 | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shimizu, A, Fukai, S, Yamagata, A, Iwamoto, A. | | Deposit date: | 2012-11-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structure of TCR and antigen complexes at an immunodominant CTL epitope in HIV-1 infection
SCI REP, 3, 2013
|
|
4OQN
 
 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with EdU | | Descriptor: | 2'-deoxy-5-ethynyluridine, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Neef, A.B, Westermaier, Y, Perozzo, R, Luedtke, N.W, Scapozza, L. | | Deposit date: | 2014-02-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with EdU
To be Published
|
|
2TSS
 
 | | TOXIC SHOCK SYNDROME TOXIN-1 FROM STAPHYLOCOCCUS AUREUS: ORTHORHOMBICC222(1) CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-04 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
2UPJ
 
 | |
4BDY
 
 | | PFV intasome with inhibitor XZ-89 | | Descriptor: | 17 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (TRANSFERRED STRAND), 19 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (NON-TRANSFERRED STRAND), 2-(3-chloro-4-fluorobenzyl)-4,5-dihydroxy-1H-isoindole-1,3(2H)-dione, ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Activities, Crystal Structures and Molecular Dynamics of Dihydro-1H-Isoindole Derivatives, Inhibitors of HIV-1 Integrase.
Acs Chem.Biol., 8, 2013
|
|
3LP6
 
 | | Crystal Structure of Rv3275c-E60A from Mycobacterium tuberculosis at 1.7A resolution | | Descriptor: | FORMIC ACID, GLYCEROL, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kim, H, Yu, M, Hung, L.-W, Terwilliger, T.C, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal Structure of Rv3275c-E60A from Mycobacterium tuberculosis at 1.7A
Resolution
To be Published
|
|
4IXD
 
 | | X-ray structure of lfa-1 i-domain in complex with ibe-667 at 1.8a resolution | | Descriptor: | 4-(3-{4-[(3-aminopropyl)carbamoyl]phenyl}-1H-indazol-1-yl)-N-methylbenzamide, Integrin alpha-L, MAGNESIUM ION | | Authors: | Kallen, J. | | Deposit date: | 2013-01-25 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and x-ray structure based investigation of an ICAM-1 binding enhancing small molecule activator of LFA-1
To be Published, 2013
|
|
4OXB
 
 | |
5NY3
 
 | | Carbonic Anhydrase II Inhibitor RA11 | | Descriptor: | 1-(4-chlorophenyl)-3-[2-(4-sulfamoylphenyl)ethyl]urea, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
4J4A
 
 | |
2DE4
 
 | | Crystal structure of DSZB C27S mutant in complex with biphenyl-2-sulfinic acid | | Descriptor: | 1,1'-BIPHENYL-2-SULFINIC ACID, ACETATE ION, DIBENZOTHIOPHENE DESULFURIZATION ENZYME B | | Authors: | Lee, W.C, Ohshiro, T, Matsubara, T, Izumi, Y, Tanokura, M. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and desulfurization mechanism of 2'-hydroxybiphenyl-2-sulfinic acid desulfinase.
J.Biol.Chem., 281, 2006
|
|
