2P1T
 
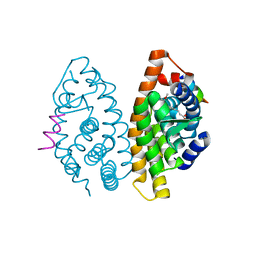 | | Crystal structure of the ligand binding domain of the retinoid X receptor alpha in complex with 3-(2'-methoxy)-tetrahydronaphtyl cinnamic acid and a fragment of the coactivator TIF-2 | | Descriptor: | (2E)-3-[4-HYDROXY-3-(3-METHOXY-5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PHENYL]ACRYLIC ACID, Nuclear receptor coactivator 2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Bourguet, W, Nahoum, V. | | Deposit date: | 2007-03-06 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7XVY
 
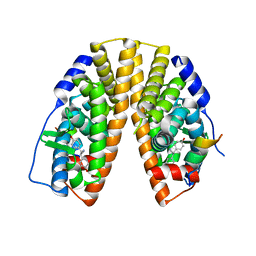 | | Human Estrogen Receptor beta Ligand-binding Domain in Complex with S-DPN | | Descriptor: | (2~{S})-2,3-bis(4-hydroxyphenyl)propanenitrile, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Furuya, N, Handa, C. | | Deposit date: | 2022-05-25 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | Evaluating the correlation of binding affinities between isothermal titration calorimetry and fragment molecular orbital method of estrogen receptor beta with diarylpropionitrile (DPN) or DPN derivatives.
J.Steroid Biochem.Mol.Biol., 222, 2022
|
|
7XWR
 
 | |
7XWQ
 
 | |
7XWP
 
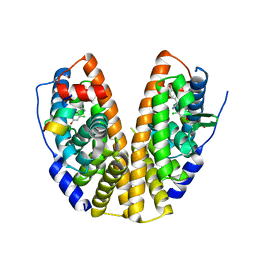 | |
3UYH
 
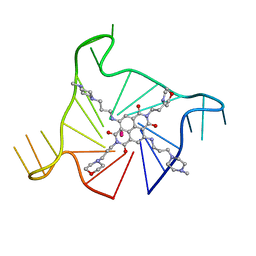 | | Crystal structure of an intramolecular human telomeric DNA G-quadruplex bound by the naphthalene diimide compound, MM41 | | Descriptor: | 4,9-bis{[3-(4-methylpiperazin-1-yl)propyl]amino}-2,7-bis[3-(morpholin-4-yl)propyl]benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, POTASSIUM ION, human telomeric DNA sequence | | Authors: | Collie, G.W, Neidle, S. | | Deposit date: | 2011-12-06 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design and evaluation of naphthalene diimide g-quadruplex ligands as telomere targeting agents in pancreatic cancer cells.
J.Med.Chem., 56, 2013
|
|
6D6B
 
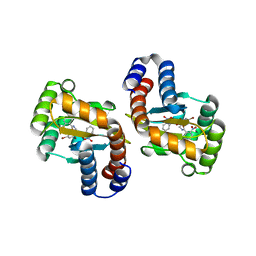 | |
7XVZ
 
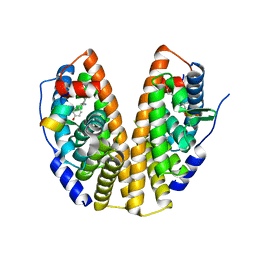 | |
2P1V
 
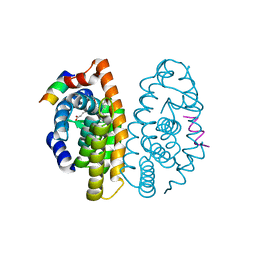 | | Crystal structure of the ligand binding domain of the retinoid X receptor alpha in complex with 3-(2'-propoxy)-tetrahydronaphtyl cinnamic acid and a fragment of the coactivator TIF-2 | | Descriptor: | (2E)-3-[4-HYDROXY-3-(5,5,8,8-TETRAMETHYL-3-PROPOXY-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)PHENYL]ACRYLIC ACID, Nuclear receptor coactivator 2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Bourguet, W, Nahoum, V. | | Deposit date: | 2007-03-06 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4MLX
 
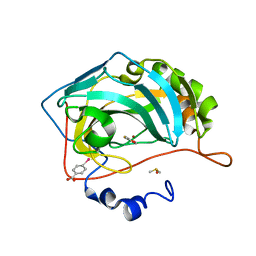 | |
5DVF
 
 | | Crystal structure of unliganded periplasmic glucose binding protein (ppGBP) from P. putida CSV86 | | Descriptor: | Binding protein component of ABC sugar transporter, SULFATE ION | | Authors: | Pandey, S, Modak, A, Phale, P.S, Bhaumik, P. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-17 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Resolution Structures of Periplasmic Glucose-binding Protein of Pseudomonas putida CSV86 Reveal Structural Basis of Its Substrate Specificity
J.Biol.Chem., 291, 2016
|
|
2FTZ
 
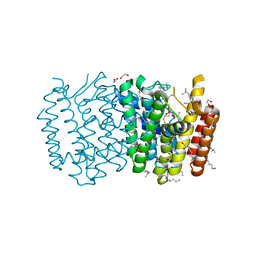 | |
8B61
 
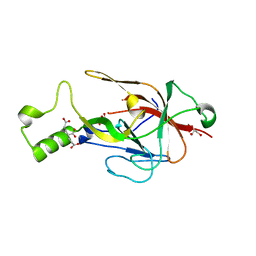 | | Crystal structure of BfrC protein from Bacteroides fragilis NCTC 9343 | | Descriptor: | Conserved hypothetical lipoprotein, GLYCEROL, pentane-1,3,5-tricarboxylic acid | | Authors: | Antonyuk, S.V, Barnett, K, Strange, R.W, Olczak, T. | | Deposit date: | 2022-09-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Bacteroides fragilis expresses three proteins similar to Porphyromonas gingivalis HmuY: Hemophore-like proteins differentially evolved to participate in heme acquisition in oral and gut microbiomes.
Faseb J., 37, 2023
|
|
8B6A
 
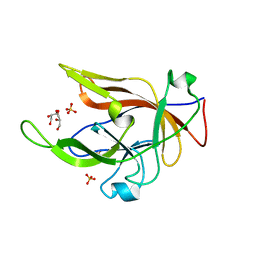 | |
8BD3
 
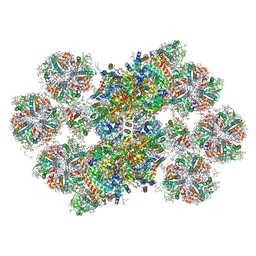 | | Cryo-EM structure of the Photosystem II - LHCII supercomplex from Chlorella ohadi | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R)-beta,beta-caroten-3-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Fadeeva, M, Klaiman, D, Caspy, I, Nelson, N. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structure of Chlorella ohadii Photosystem II Reveals Protective Mechanisms against Environmental Stress.
Cells, 12, 2023
|
|
7K42
 
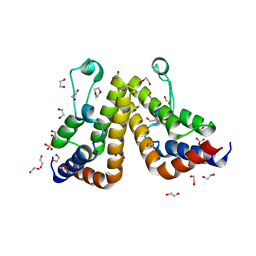 | | Crystal structure of the second bromodomain (BD2) of human TAF1 bound to dioxane | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, SULFATE ION, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
4LZ7
 
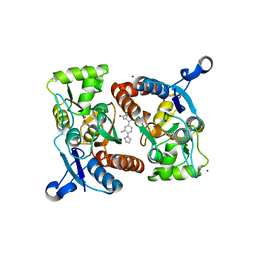 | |
5N21
 
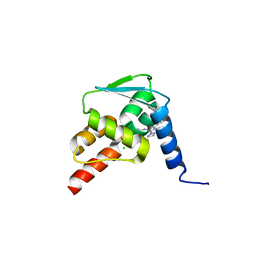 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | 2-[(2~{S})-1-[3-cyano-7-[(2-oxidanylidene-3,4-dihydro-1~{H}-quinolin-6-yl)amino]pyrazolo[1,5-a]pyrimidin-5-yl]pyrrolidin-2-yl]ethanoic acid, B-cell lymphoma 6 protein, CHLORIDE ION | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
8FPC
 
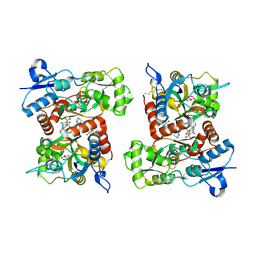 | | Conformation 1 of the ligand binding domain (LBDconf1) of GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg) | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-04 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FPH
 
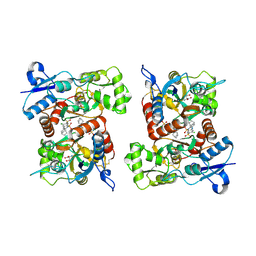 | | Conformation 2 of the ligand binding domain (LBDconf2) of GluA2 flip Q isoform of AMPA receptor in complex with gain-of-function TARP gamma-2, with 10mM CaCl2, 150mM NaCl, 1mM MgCl2, 330uM CTZ, and 100mM glutamate (Open-CaNaMg) | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Nakagawa, T. | | Deposit date: | 2023-01-04 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The open gate of the AMPA receptor forms a Ca 2+ binding site critical in regulating ion transport.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5N1X
 
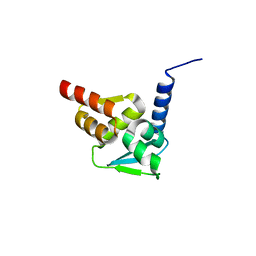 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-ethyl-5-pyridin-3-yl-pyrazolo[1,5-a]pyrimidin-7-amine | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N1Z
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine macrocyclic ligand | | Descriptor: | B-cell lymphoma 6 protein, CHLORIDE ION, pyrazolo-pyrimidine macrocycle | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
3IT2
 
 | | Crystal structure of ligand-free Francisella tularensis histidine acid phosphatase | | Descriptor: | ACETATE ION, Acid phosphatase | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
5N1V
 
 | | Crystal structure of the protein kinase CK2 catalytic subunit in complex with pyrazolo-pyrimidine macrocyclic ligand | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N20
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-[5-[[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]amino]-2-[(3~{R})-3-(dimethylamino)pyrrolidin-1-yl]phenyl]ethanamide | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
