6D74
 
 | |
6O1A
 
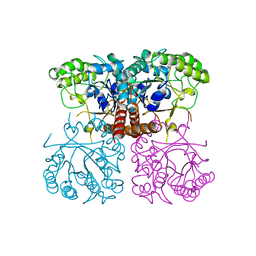 | |
6NMI
 
 | | Cryo-EM structure of the human TFIIH core complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription and DNA repair factor IIH helicase subunit XPB, General transcription and DNA repair factor IIH helicase subunit XPD, ... | | Authors: | Greber, B.J, Toso, D, Fang, J, Nogales, E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-03-13 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The complete structure of the human TFIIH core complex.
Elife, 8, 2019
|
|
5CLV
 
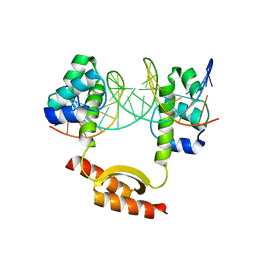 | | Crystal Structure of KorA-operator DNA complex (KorA-OA) | | Descriptor: | 5'-D(CP*CP*AP*AP*GP*TP*TP*TP*AP*GP*CP*TP*AP*AP*AP*CP*TP*TP*GP*GP*)-3', TrfB transcriptional repressor protein | | Authors: | White, S.A, Hyde, E.I, Rajasekar, K.V. | | Deposit date: | 2015-07-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
6NS2
 
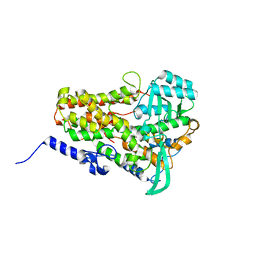 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. P212121 crystal form. | | Descriptor: | FE (II) ION, lipoxygenase | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
7FIH
 
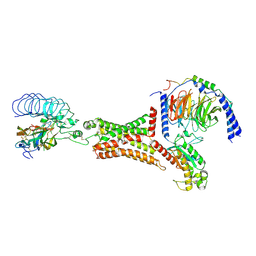 | | luteinizing hormone/choriogonadotropin receptor(S277I)-chorionic gonadotropin-Gs-Org43553 complex | | Descriptor: | 5-azanyl-N-tert-butyl-2-methylsulfanyl-4-[3-(2-morpholin-4-ylethanoylamino)phenyl]thieno[2,3-d]pyrimidine-6-carboxamide, Choriogonadotropin subunit beta 3, Engineered Guanine nucleotide-binding protein G(s) subunit alpha, ... | | Authors: | Duan, J, Xu, P, Cheng, X, Mao, C, Croll, T, He, X, Shi, J, Luan, X, Yin, W, You, E, Liu, Q, Zhang, S, Jiang, H, Zhang, Y, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-07-31 | | Release date: | 2021-09-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of full-length glycoprotein hormone receptor signalling complexes.
Nature, 598, 2021
|
|
7M99
 
 | | ATPgS bound TnsC filament from ShCAST system | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Park, J, Tsai, A.W.L, Mehrotra, E, Kellogg, E.H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for target site selection in RNA-guided DNA transposition systems.
Science, 373, 2021
|
|
2H5U
 
 | | Crystal structure of laccase from Cerrena maxima at 1.9A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lyashenko, A.V, Gabdoulkhakov, A.G, Zaitsev, V.N, Lamzin, V.S, Lindley, P.F, Bento, I, Betzel, C, Zhukhlistova, N.E, Zhukova, Y.N, Mikhailov, A.M. | | Deposit date: | 2006-05-27 | | Release date: | 2007-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Purification, crystallization and preliminary X-ray study of the fungal laccase from Cerrena maxima
Acta Crystallogr.,Sect.F, 62, 2006
|
|
7M9A
 
 | | ADP-AlF3 bound TnsC structure from ShCAST system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (27-MER), TnsC | | Authors: | Park, J, Tsai, A.W.L, Mehrotra, E, Kellogg, E.H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for target site selection in RNA-guided DNA transposition systems.
Science, 373, 2021
|
|
7M9C
 
 | | ADP-AlF3 bound TnsC structure in open form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (34-MER), TnsC | | Authors: | Park, J, Tsai, A.W.L, Mehrotra, E, Kellogg, E.H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for target site selection in RNA-guided DNA transposition systems.
Science, 373, 2021
|
|
7M9B
 
 | | ADP-AlF3 bound TnsC structure in closed form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (27-MER), TnsC | | Authors: | Park, J, Tsai, A.W.L, Mehrotra, E, Kellogg, E.H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for target site selection in RNA-guided DNA transposition systems.
Science, 373, 2021
|
|
6NZ1
 
 | |
7MRU
 
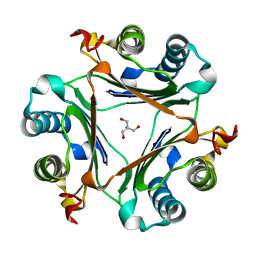 | | Crystal structure of S62A MIF2 mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-dopachrome decarboxylase | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
6NS4
 
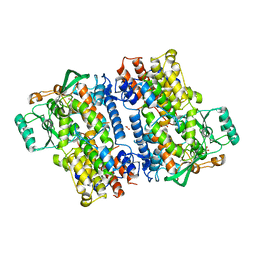 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. C2 crystal form. | | Descriptor: | ACETATE ION, FE (II) ION, GLYCEROL, ... | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
2XR9
 
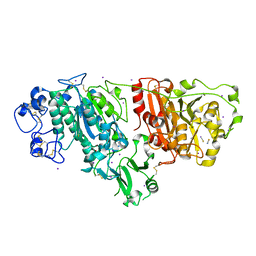 | | Crystal structure of Autotaxin (ENPP2) | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Kamtekar, S, Hausmann, J, Day, J.E, Christodoulou, E, Perrakis, A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate discrimination and integrin binding by autotaxin.
Nat. Struct. Mol. Biol., 18, 2011
|
|
4CFF
 
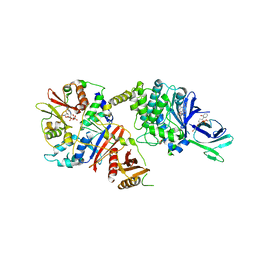 | | Structure of full length human AMPK in complex with a small molecule activator, a thienopyridone derivative (A-769662) | | Descriptor: | 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Carmena, D, Bright, N.J, Haire, L.F, Underwood, E, Patel, B.R, Heath, R.B, Walker, P.A, Hallen, S, Giordanetto, F, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.924 Å) | | Cite: | Structural Basis of Ampk Regulation by Small Molecule Activators.
Nat.Commun., 4, 2013
|
|
1LP9
 
 | | Xenoreactive complex AHIII 12.2 TCR bound to p1049/HLA-A2.1 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Buslepp, J, Wang, H, Biddison, W.E, Appella, E, Collins, E.J. | | Deposit date: | 2002-05-07 | | Release date: | 2003-11-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A correlation between TCR Valpha docking on MHC and CD8 dependence: implications for T cell selection.
Immunity, 19, 2003
|
|
7MSE
 
 | | High-resolution crystal structure of hMIF2 with tartrate at the active site | | Descriptor: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
4NWP
 
 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage, T33-21, Crystallized in Space Group R32 | | Descriptor: | AMMONIUM ION, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
7JJM
 
 | | Crystal structure of Importin alpha 2 in complex with LSD1 NLS | | Descriptor: | CHLORIDE ION, Importin subunit alpha-1, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McGuaig, R, Tan, H.Y.A, Hardy, C, Seddiki, N, Ali, S, Dahlstrom, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K.M, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
7JK7
 
 | | Crystal structure of Importin alpha 2 in complex with LSD1 NLS S111E mutant | | Descriptor: | GLYCEROL, Importin subunit alpha-1, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McCuaig, R, Tan, A.H.Y, Hardy, K, Seddiki, N, Ali, S, Dahlstron, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K.M, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
5E9Z
 
 | | Cytochrome P450 BM3 mutant M11 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Bifunctional cytochrome P450/NADPH--P450 reductase, FE (II) ION, ... | | Authors: | Capoferri, L, Leth, R, ter Haar, E, Mohanty, A.K, Grootenhuis, D.J, Vottero, E, Commandeur, J.N.M, Vermeulen, N.P.E, Jorgensen, F.S, Olsen, L, Geerke, D.P. | | Deposit date: | 2015-10-15 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights into regioselective metabolism of mefenamic acid by cytochrome P450 BM3 mutants through crystallography, docking, molecular dynamics, and free energy calculations.
Proteins, 84, 2016
|
|
1YZ9
 
 | | Crystal structure of RNase III mutant E110Q from Aquifex aeolicus complexed with double stranded RNA at 2.1-Angstrom Resolution | | Descriptor: | 5'-R(*CP*GP*AP*AP*CP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III, SULFATE ION | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
2PYP
 
 | | PHOTOACTIVE YELLOW PROTEIN, PHOTOSTATIONARY STATE, 50% GROUND STATE, 50% BLEACHED | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Genick, U.K, Borgstahl, G.E.O, Ng, K, Ren, Z, Pradervand, C, Burke, P, Srajer, V, Teng, T, Schildkamp, W, Mcree, D.E, Moffat, K, Getzoff, E.D. | | Deposit date: | 1997-02-03 | | Release date: | 1998-04-29 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a protein photocycle intermediate by millisecond time-resolved crystallography.
Science, 275, 1997
|
|
7ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
