2BUD
 
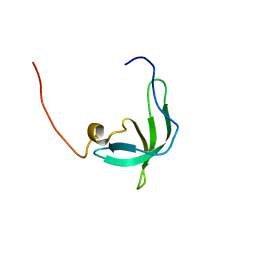 | | The solution structure of the chromo barrel domain from the males- absent on the first (MOF) protein | | Descriptor: | MALES-ABSENT ON THE FIRST PROTEIN | | Authors: | Nielsen, P.R, Nietlispach, D, Buscaino, A, Warner, R.J, Akhtar, A, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2005-06-09 | | Release date: | 2005-06-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Chromo Barrel Domain from the Mof Acetyltransferase
J.Biol.Chem., 280, 2005
|
|
2BVB
 
 | | The C-terminal domain from Micronemal Protein 1 (MIC1) from Toxoplasma Gondii | | Descriptor: | MICRONEMAL PROTEIN 1 | | Authors: | Saouros, S, Edwards-Jones, B, Reiss, M, Sawmynaden, K, Cota, E, Simpson, P, Dowse, T.J, Jakle, U, Ramboarina, S, Shivarattan, T, Matthews, S, Soldati-Favre, D. | | Deposit date: | 2005-06-23 | | Release date: | 2005-10-12 | | Last modified: | 2021-06-23 | | Method: | SOLUTION NMR | | Cite: | A Novel Galectin-Like Domain from Toxoplasma Gondll Micronemal Protein 1 Assists the Folding, Assembly,and Transport of a Cell-Adhesion Complex.
J.Biol.Chem., 280, 2005
|
|
2AME
 
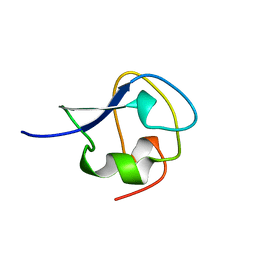 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14Q | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-18 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
2ARW
 
 | | The solution structure of the membrane proximal cytokine receptor domain of the human interleukin-6 receptor | | Descriptor: | Interleukin-6 receptor alpha chain | | Authors: | Hecht, O, Dingley, A.J, Schwantner, A, Ozbek, S, Rose-John, S, Grotzinger, J. | | Deposit date: | 2005-08-22 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the membrane-proximal cytokine receptor domain of the human interleukin-6 receptor
Biol.Chem., 387, 2006
|
|
2DYD
 
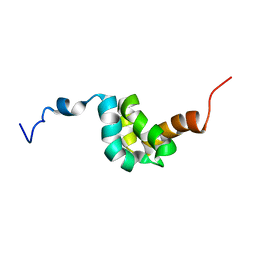 | |
2AP0
 
 | |
2AWQ
 
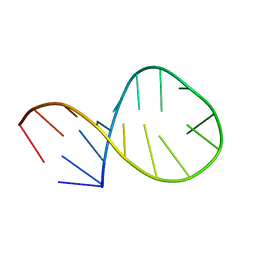 | |
2AIV
 
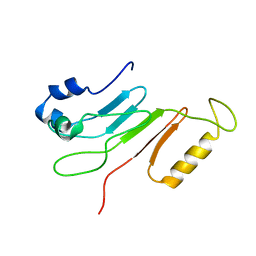 | | Multiple conformations in the ligand-binding site of the yeast nuclear pore targeting domain of NUP116P | | Descriptor: | fragment of Nucleoporin NUP116/NSP116 | | Authors: | Robinson, M.A, Park, S, Sun, Z.-Y.J, Silver, P, Wagner, G, Hogle, J. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple Conformations in the Ligand-binding Site of the Yeast Nuclear Pore-targeting Domain of Nup116p
J.Biol.Chem., 280, 2005
|
|
2EQH
 
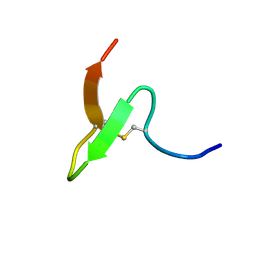 | | Solution structure of growth-blocking peptide of the armyworm, Pseudaletia separata | | Descriptor: | Growth-blocking peptide, short form | | Authors: | Umetsu, Y, Aizawa, T, Kamiya, M, Kumaki, Y, Demura, M, Kawano, K. | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-01 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | C-terminal elongation of growth-blocking peptide enhances its biological activity and micelle binding affinity
J.Biol.Chem., 284, 2009
|
|
2EQT
 
 | | Micelle-bound structure of growth-blocking peptide of the armyworm, Pseudaletia separata | | Descriptor: | Growth-blocking peptide, long form | | Authors: | Umetsu, Y, Aizawa, T, Kamiya, M, Kumaki, Y, Demura, M, Kawano, K. | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-01 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | C-terminal elongation of growth-blocking peptide enhances its biological activity and micelle binding affinity
J.Biol.Chem., 284, 2009
|
|
2ERL
 
 | | PHEROMONE ER-1 FROM | | Descriptor: | ETHANOL, MATING PHEROMONE ER-1 | | Authors: | Anderson, D.H, Weiss, M.S, Eisenberg, D. | | Deposit date: | 1995-12-20 | | Release date: | 1996-07-11 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A challenging case for protein crystal structure determination: the mating pheromone Er-1 from Euplotes raikovi.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2B6G
 
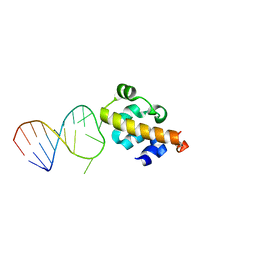 | |
2B7G
 
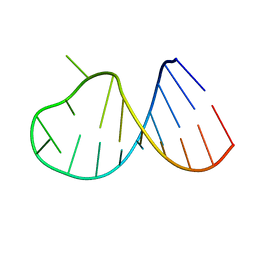 | |
2AP8
 
 | |
2D49
 
 | | Solution structure of the Chitin-Binding Domain of Streptomyces griseus Chitinase C | | Descriptor: | chitinase C | | Authors: | Akagi, K, Watanabe, J, Hara, M, Kezuka, Y, Chikaishi, E, Yamaguchi, T, Akutsu, H, Nonaka, T, Watanabe, T, Ikegami, T. | | Deposit date: | 2005-10-11 | | Release date: | 2006-10-11 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Identification of the substrate interaction region of the chitin-binding domain of Streptomyces griseus chitinase C
J.Biochem.(Tokyo), 139, 2006
|
|
2DA8
 
 | | SOLUTION STRUCTURE OF A COMPLEX BETWEEN (N-MECYS3,N-MECYS7)TANDEM AND (D(GATATC))2 | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(*GP*AP*TP*AP*TP*C)-3'), TRIOSTIN A | | Authors: | Addess, K.J, Sinsheimer, J.S, Feigon, J. | | Deposit date: | 1993-04-09 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Complex between [N-Mecys3,N-Mecys7]Tandem and [D(Gatatc)]2.
Biochemistry, 32, 1993
|
|
2DVJ
 
 | | phosphorylated Crk-II | | Descriptor: | V-crk sarcoma virus CT10 oncogene homolog, isoform a | | Authors: | Kobashigawa, Y, Inagaki, F. | | Deposit date: | 2006-07-31 | | Release date: | 2007-05-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2DRN
 
 | | Docking and dimerization domain (D/D) of the Type II-alpha regulatory subunity of protein kinase A (PKA) in complex with a peptide from an A-kinase anchoring protein | | Descriptor: | 24-residues peptide from an a-kinase anchoring protein, cAMP-dependent protein kinase type II-alpha regulatory subunit | | Authors: | Newlon, M.G, Roy, M, Morikis, D, Hausken, Z.E, Coghlan, V, Scott, J.D, Jennings, P.A. | | Deposit date: | 2006-06-11 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes.
Embo J., 20, 2001
|
|
2DTB
 
 | |
2FJL
 
 | | Solution Structure of the Split PH domain in Phospholipase C-gamma1 | | Descriptor: | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2006-01-03 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of the Split Pleckstrin Homology Domain in Phospholipase C-{gamma}1 and Its Interaction with TRPC3
J.Biol.Chem., 281, 2006
|
|
2ERS
 
 | |
2CKC
 
 | | Solution structures of the BRK domains of the human Chromo Helicase Domain 7 and 8, reveals structural similarity with GYF domain suggesting a role in protein interaction | | Descriptor: | CHROMODOMAIN-HELICASE-DNA-BINDING PROTEIN 7 | | Authors: | Ab, E, de Jong, R.N, Diercks, T, Xiaoyun, J, Daniels, M, Kaptein, R, Folkers, G.E. | | Deposit date: | 2006-04-14 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Brk Domains of the Human Chromo Helicase Domain 7 and 8, Reveals Structural Similarity with Gyf Domain Suggesting a Role in Protein Interaction
To be Published
|
|
2CTI
 
 | |
2FLH
 
 | | Crystal structure of cytokinin-specific binding protein from mung bean in complex with cytokinin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, SODIUM ION, cytokinin-specific binding protein | | Authors: | Pasternak, O, Bujacz, G.D, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Vigna radiata Cytokinin-Specific Binding Protein in Complex with Zeatin.
Plant Cell, 18, 2006
|
|
2EFF
 
 | |
