7DRT
 
 | | Human Wntless in complex with Wnt3a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhong, Q, Zhao, Y, Ye, F, Xiao, Z, Huang, G, Zhang, Y, Lu, P, Xu, W, Zhou, Q, Ma, D. | | Deposit date: | 2020-12-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structure of human Wntless in complex with Wnt3a.
Nat Commun, 12, 2021
|
|
4GC6
 
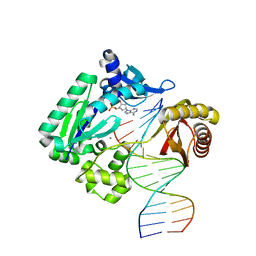 | | Crystal structure of Dpo4 in complex with N-MC-dAMP opposite dT | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), DNA (5'-D(*TP*CP*AP*TP*GP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Eoff, R.L, Ketkar, A, Banerjee, S, Zafar, M.K. | | Deposit date: | 2012-07-29 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Differential furanose selection in the active sites of archaeal DNA polymerases probed by fixed-conformation nucleotide analogues.
Biochemistry, 51, 2012
|
|
4FZJ
 
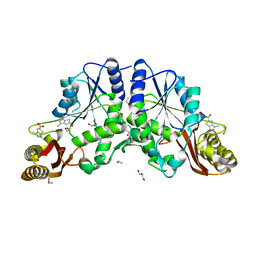 | | Pantothenate synthetase in complex with 1,3-DIMETHYL-1H-THIENO[2,3-C]PYRAZOLE-5-CARBOXYLIC ACID | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-1H-thieno[2,3-c]pyrazole-5-carboxylic acid, ETHANOL, ... | | Authors: | Silvestre, H.L, Blundell, T.L, Abell, C, Ciulli, A. | | Deposit date: | 2012-07-06 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7ZC7
 
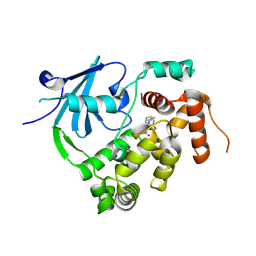 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012941 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012941
To Be Published
|
|
4FZZ
 
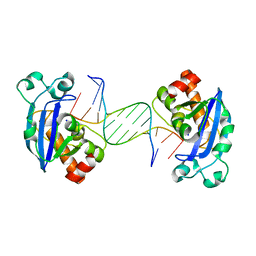 | | Exonuclease X in complex with 5' overhanging duplex DNA | | Descriptor: | DNA (5'-D(*GP*TP*CP*AP*TP*TP*GP*TP*GP*GP*AP*TP*CP*CP*GP*AP*G)-3'), Exodeoxyribonuclease 10, SODIUM ION | | Authors: | Wang, T, Sun, H, Cheng, F, Bi, L, Jiang, T. | | Deposit date: | 2012-07-08 | | Release date: | 2013-07-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition and processing of double-stranded DNA by ExoX, a distributive 3'-5' exonuclease
Nucleic Acids Res., 41, 2013
|
|
5I4O
 
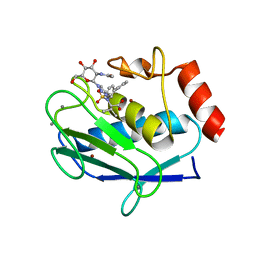 | | Crystal structure of the catalytic domain of MMP-12 in complex with a selective sugar-conjugated triazole-linked carboxylate zinc-chelator water-soluble inhibitor (DC28). | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-({1-[2-(acetylamino)-2-deoxy-beta-D-glucopyranosyl]-1H-1,2,3-triazol-4-yl}methyl)-N-[([1,1'-biphenyl]-4-yl)sulfonyl]-D-valine, ... | | Authors: | Stura, E.A, Rosalia, L, Cuffaro, D, Tepshi, L, Ciccone, L, Rossello, A. | | Deposit date: | 2016-02-12 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors.
Chemmedchem, 11, 2016
|
|
2GFE
 
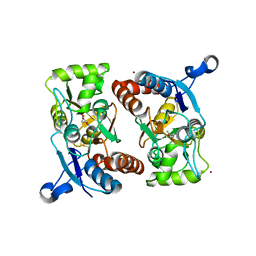 | |
4HQN
 
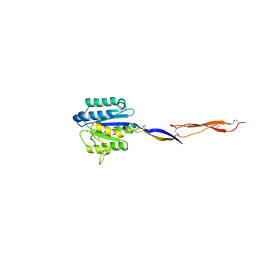 | | Crystal structure of manganese-loaded Plasmodium vivax TRAP protein | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Sporozoite surface protein 2, ... | | Authors: | Song, G, Koksal, A.C, Lu, C, Springer, T.A. | | Deposit date: | 2012-10-25 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Shape change in the receptor for gliding motility in Plasmodium sporozoites.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7YW3
 
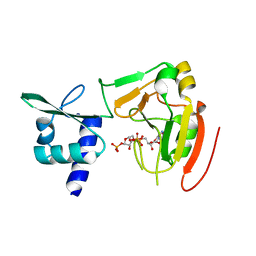 | | Crystal structure of tRNA 2'-phosphotransferase from Homo sapiens | | Descriptor: | 1,2-ETHANEDIOL, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate, tRNA 2'-phosphotransferase 1 | | Authors: | Yang, X.Y, Liu, X.H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical insights into the molecular mechanism of TRPT1 for nucleic acid ADP-ribosylation.
Nucleic Acids Res., 51, 2023
|
|
7YW4
 
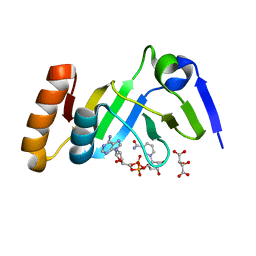 | |
2GAG
 
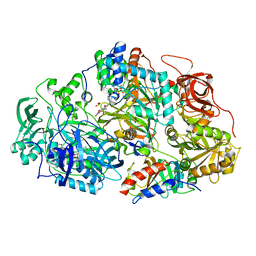 | | Heteroteterameric sarcosine: structure of a diflavin metaloenzyme at 1.85 a resolution | | Descriptor: | 2-FUROIC ACID, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Chen, Z.W, Hassan-Abdulah, A, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Heterotetrameric sarcosine oxidase: structure of a diflavin metalloenzyme at 1.85 a resolution.
J.Mol.Biol., 360, 2006
|
|
7ZCB
 
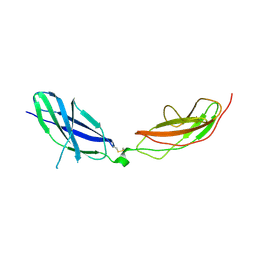 | | Human Pikachurin/EGFLAM N-terminal Fibronectin-III (1-2) domains | | Descriptor: | CHLORIDE ION, Pikachurin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Pantalone, S, Savino, S, Viti, L.V, Forneris, F. | | Deposit date: | 2022-03-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the photoreceptor synaptic assembly of the extracellular matrix protein pikachurin with the orphan receptor GPR179.
Sci.Signal., 16, 2023
|
|
6LXN
 
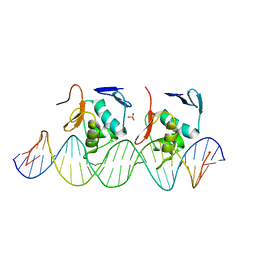 | |
2GB8
 
 | | Solution structure of the complex between yeast iso-1-cytochrome c and yeast cytochrome c peroxidase | | Descriptor: | Cytochrome c iso-1, Cytochrome c peroxidase, HEME C, ... | | Authors: | Volkov, A.N, Worrall, J.A.R, Ubbink, M. | | Deposit date: | 2006-03-10 | | Release date: | 2006-11-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5I23
 
 | | Crystal Structure of Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31, in complex with Cyclophellitol Aziridine probe CF022 | | Descriptor: | 1,2-ETHANEDIOL, 1-(8-{[(1S,2R,3S,4S,5R,6R)-2,3,4,5-tetrahydroxy-6-(hydroxymethyl)cyclohexyl]amino}octyl)triaza-1,2-dien-2-ium, Oligosaccharide 4-alpha-D-glucosyltransferase, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2016-02-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detection of Active Mammalian GH31 alpha-Glucosidases in Health and Disease Using In-Class, Broad-Spectrum Activity-Based Probes.
Acs Cent.Sci., 2, 2016
|
|
6LYJ
 
 | | The crystal structure of SAUGI/EBVUDG complex | | Descriptor: | SAUGI, Uracil-DNA glycosylase | | Authors: | Liao, Y.T, Ko, T.P, Wang, H.C. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the differential interactions between the DNA mimic protein SAUGI and two gamma herpesvirus uracil-DNA glycosylases.
Int.J.Biol.Macromol., 160, 2020
|
|
7YW2
 
 | | Crystal structure of tRNA 2'-phosphotransferase from Mus musculus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, GLYCINE, ... | | Authors: | Yang, X.Y, Liu, X.H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical insights into the molecular mechanism of TRPT1 for nucleic acid ADP-ribosylation.
Nucleic Acids Res., 51, 2023
|
|
2G2B
 
 | | NMR structure of the human allograft inflammatory factor 1 | | Descriptor: | Allograft inflammatory factor 1 | | Authors: | Song, J, Tyler, R.C, Newman, C.L, Vinarov, D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-15 | | Release date: | 2006-02-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human allograft inflammatory factor 1
To be published
|
|
7YNQ
 
 | |
2GE5
 
 | |
7DEG
 
 | | Cryo-EM structure of a heme-copper terminal oxidase dimer provides insights into its catalytic mechanism | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guoliang, Z, Shuangbo, Z. | | Deposit date: | 2020-11-04 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Unusual Homodimer of a Heme-Copper Terminal Oxidase Allows Itself to Utilize Two Electron Donors.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6LPC
 
 | | Crystal Structure of rat Munc18-1 with K332E/K333E mutation | | Descriptor: | Syntaxin-binding protein 1 | | Authors: | Wang, X.P, Gong, J.H, Wang, S, Zhu, L, Yang, X.Y, Xu, Y.Y, Yang, X.F, Ma, C. | | Deposit date: | 2020-01-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | Munc13 activates the Munc18-1/syntaxin-1 complex and enables Munc18-1 to prime SNARE assembly.
Embo J., 39, 2020
|
|
7YNT
 
 | |
4L5I
 
 | |
7YNM
 
 | | ThT-bound alpha-synuclein fibrils conformation 1 | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, Alpha-synuclein | | Authors: | Tao, Y.Q, Zhao, Q.Y, Liu, C, Li, D. | | Deposit date: | 2022-07-31 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | ThT-bound alpha-synuclein fibrils conformation 1
To Be Published
|
|
