8T89
 
 | |
8T86
 
 | |
8T85
 
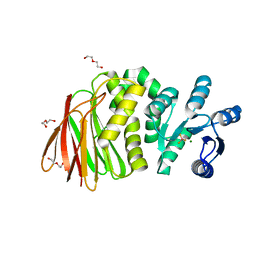 | | Structure of RssB bound to beryllofluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Brugger, C, Schwartz, J, Deaconescu, A.M. | | Deposit date: | 2023-06-21 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of phosphorylated-like RssB, the adaptor delivering sigma s to the ClpXP proteolytic machinery, reveals an interface switch for activation.
J.Biol.Chem., 299, 2023
|
|
8T84
 
 | | Racemic mixture of amyloid beta segment 35-MVGGVV-40 forms heterochiral rippled beta-sheet, includes hexafluoroisopropanol | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, Racemic mixture of amyloid beta segment 35-MVGGVV-40 | | Authors: | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Racemic Peptides from Amyloid beta and Amylin Form Rippled beta-Sheets Rather Than Pleated beta-Sheets.
J.Am.Chem.Soc., 145, 2023
|
|
8T83
 
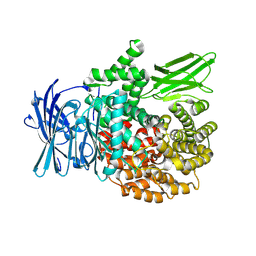 | |
8T82
 
 | | Racemic mixture of amyloid beta segment 35-MVGGVV-40 forms heterochiral rippled beta-sheet, includes pentafluoropropionic acid | | Descriptor: | amyloid beta segment 35-MVGGVV-40, racemic mixture, pentafluoropropanoic acid | | Authors: | Sawaya, M.R, Raskatov, J.A, Hazari, A. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Racemic Peptides from Amyloid beta and Amylin Form Rippled beta-Sheets Rather Than Pleated beta-Sheets.
J.Am.Chem.Soc., 145, 2023
|
|
8T81
 
 | |
8T80
 
 | | Sequence specific (AATT) orientation of Hoechst molecules at two unique minor groove binding sites within a self-assembled 3D DNA lattice (4x5) | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*GP*AP*GP*AP*AP*TP*TP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*TP*TP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-21 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8T7X
 
 | | Sequence specific (AATT) orientation of DAPI molecules at two unique minor groove binding sites within a self-assembled 3D DNA lattice (4x5) | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, CACODYLATE ION, DNA (5'-D(*GP*AP*GP*AP*AP*TP*TP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*AP*TP*TP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-21 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8T7P
 
 | | X-ray crystal structure of PfA-M1(M462S) | | Descriptor: | Aminopeptidase N, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Yang, W, Drinkwater, N, Webb, C.T, McGowan, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational dynamics of the Plasmodium falciparum M1 aminopeptidase.
To Be Published
|
|
8T7I
 
 | | Structure of the S1CE variant of Fab F1 (FabS1CE-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7B
 
 | | Sequence specific (AATT) orientation of netropsin molecules at a unique minor groove binding site (position2) within a self-assembled 3D DNA lattice (4x5) | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*TP*TP*G)-3'), DNA (5'-D(P*AP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8T70
 
 | |
8T6Z
 
 | |
8T6I
 
 | | Structure of VHH-Fab complex with engineered Crystal Kappa region | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Filippova, E.V, Thompson, I, Kossiakoff, A.A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T6H
 
 | |
8T6B
 
 | | Human VMAT2 in complex with serotonin | | Descriptor: | SEROTONIN, Synaptic vesicular amine transporter | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8T6A
 
 | | Human VMAT2 in complex with reserpine | | Descriptor: | Synaptic vesicular amine transporter, reserpine | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8T69
 
 | | Human VMAT2 in complex with tetrabenazine | | Descriptor: | Synaptic vesicular amine transporter, tetrabenazine | | Authors: | Pidathala, S, Dai, Y, Lee, C.H. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanisms of neurotransmitter transport and drug inhibition in human VMAT2.
Nature, 623, 2023
|
|
8T63
 
 | |
8T62
 
 | |
8T61
 
 | |
8T5W
 
 | |
8T5Q
 
 | |
8T5P
 
 | |
