3TQM
 
 | | Structure of an ribosomal subunit interface protein from Coxiella burnetii | | Descriptor: | Ribosome-associated factor Y, SULFATE ION | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
4COE
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl {(2S)-1-[2-(biphenyl-4-ylmethyl)-2-{(4R)-4-hydroxy-5-{[(2S)-3-methyl-1-oxo-1-(prop-2-en-1-ylamino)butan-2-yl]amino}-5-oxo-4-[4-(prop-2-en-1-yl)benzyl]pentyl}hydrazinyl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-01-28 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4CPU
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[[4-[(3S)-3,4-dihydrothiophen-3-yl]phenyl]methyl]-2-[3-[(3Z,8S,11R)-11-oxidanyl-7,10-bis(oxidanylidene)-8-propan-2-yl-6,9-diazabicyclo[11.2.2]heptadeca-1(16),3,13(17),14-tetraen-11-yl]propyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4CPX
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl ~{N}-[(2~{S})-3,3-dimethyl-1-[2-[3-[(3~{R},6~{S},10~{Z})-3-oxidanyl-4,7-bis(oxidanylidene)-6-propan-2-yl-5,8-diazabicyclo[11.2.2]heptadeca-1(16),10,13(17),14-tetraen-3-yl]propyl]-2-[(4-thiophen-2-ylphenyl)methyl]hydrazinyl]-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of P1'-functionalized macrocyclic transition-state mimicking HIV-1 protease inhibitors encompassing a tertiary alcohol.
J. Med. Chem., 57, 2014
|
|
4CPW
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[[4-[(3S)-3,4-dihydrothiophen-3-yl]phenyl]methyl]-2-[3-[(3Z,8S,11R)-11-oxidanyl-7,10-bis(oxidanylidene)-8-propan-2-yl-6,9-diazabicyclo[11.2.2]heptadeca-1(16),3,13(17),14-tetraen-11-yl]propyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4CPS
 
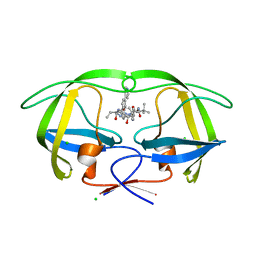 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl [(2S)-1-(2-{3-[(3R,6S,10Z)-3-hydroxy-4,7-dioxo-6-(propan-2-yl)-5,8-diazabicyclo[11.2.2]heptadeca-1(15),10,13,16-tetraen-3-yl]propyl}-2-[4-(pyridin-3-yl)benzyl]hydrazinyl)-3,3-dimethyl-1-oxobutan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4CPR
 
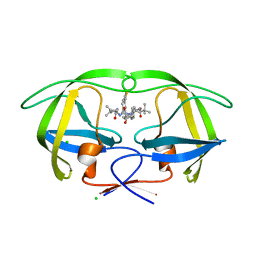 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl ~{N}-[(2~{S})-3,3-dimethyl-1-[2-[(4~{R})-5-[[(2~{S})-3-methyl-1-oxidanylidene-1-(prop-2-enylamino)butan-2-yl]amino]-4-oxidanyl-5-oxidanylidene-4-[(4-prop-2-enylphenyl)methyl]pentyl]-2-[(4-thiophen-2-ylphenyl)methyl]hydrazinyl]-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
4CP7
 
 | | Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol | | Descriptor: | CHLORIDE ION, PROTEASE, methyl [(2S)-1-(2-{(4R)-4-hydroxy-5-{[(2S)-3-methyl-1-oxo-1-(prop-2-en-1-ylamino)butan-2-yl]amino}-5-oxo-4-[4-(prop-2-en-1-yl)benzyl]pentyl}-2-[4-(pyridin-4-yl)benzyl]hydrazinyl)-3,3-dimethyl-1-oxobutan-2-yl]carbamate | | Authors: | deRosa, M, Unge, J, Motwani, H.V, Rosenquist, A, Vrang, L, Wallberg, H, Larhed, M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of P1'-Functionalized Macrocyclic Transition-State Mimicking HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol.
J.Med.Chem., 57, 2014
|
|
5C9H
 
 | | Structural Basis of Template Boundary Definition in Tetrahymena Telomerase | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*GP*AP*AP*CP*UP*GP*UP*CP*A)-3'), RNA (5'-R(P*UP*CP*AP*UP*UP*CP*AP*GP*UP*UP*CP*U)-3'), ... | | Authors: | Jansson, L.I, Akiyama, B.M, Ooms, A, Lu, C, Rubin, S.M, Stone, M.D. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of template-boundary definition in Tetrahymena telomerase.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5MDH
 
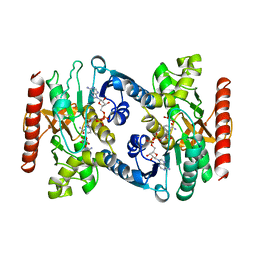 | | CRYSTAL STRUCTURE OF TERNARY COMPLEX OF PORCINE CYTOPLASMIC MALATE DEHYDROGENASE ALPHA-KETOMALONATE AND TNAD AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-KETOMALONIC ACID, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chapman, A.D.M, Cortes, A, Dafforn, T.R, Clarke, A.R, Brady, R.L. | | Deposit date: | 1998-10-08 | | Release date: | 1999-05-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of substrate specificity in malate dehydrogenases: crystal structure of a ternary complex of porcine cytoplasmic malate dehydrogenase, alpha-ketomalonate and tetrahydoNAD.
J.Mol.Biol., 285, 1999
|
|
3CHV
 
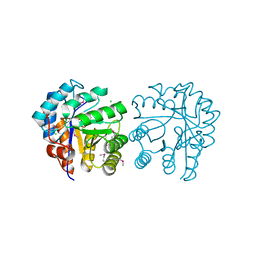 | |
3CZR
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) in Complex with Arylsulfonylpiperazine Inhibitor | | Descriptor: | (2R)-1-[(4-tert-butylphenyl)sulfonyl]-2-methyl-4-(4-nitrophenyl)piperazine, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Corticosteroid 11-beta-dehydrogenase isozyme 1, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2008-04-29 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and Initial SAR of Arylsulfonylpiperazine Inhibitors of 11beta-Hydroxysteroid Dehydrogenase Type 1 (11beta-HSD1)
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6X2A
 
 | |
6X29
 
 | |
5K6D
 
 | | Structure of FS50 an antagonist of NaV1.5 | | Descriptor: | Putative secreted salivary protein | | Authors: | Andersen, J.F, Xu, X. | | Deposit date: | 2016-05-24 | | Release date: | 2016-11-23 | | Method: | X-RAY DIFFRACTION (1.139 Å) | | Cite: | Structure and Function of FS50, a salivary protein from the flea Xenopsylla cheopis that blocks the sodium channel NaV1.5.
Sci Rep, 6, 2016
|
|
6T8Y
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
8DGF
 
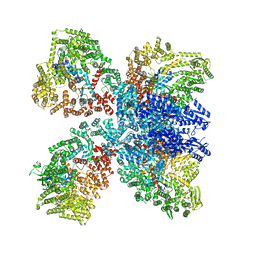 | | Avs4 bound to phage PhiV-1 portal | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding protein Avs4, MAGNESIUM ION, ... | | Authors: | Wilkinson, M.E, Gao, L, Strecker, J, Makarova, K.S, Macrae, R.K, Koonin, E.V, Zhang, F. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Prokaryotic innate immunity through pattern recognition of conserved viral proteins.
Science, 377, 2022
|
|
8DGC
 
 | | Avs3 bound to phage PhiV-1 terminase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SeAvs3, ... | | Authors: | Wilkinson, M.E, Gao, L, Strecker, J, Makarova, K.S, Macrae, R.K, Koonin, E.V, Zhang, F. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Prokaryotic innate immunity through pattern recognition of conserved viral proteins.
Science, 377, 2022
|
|
6X2B
 
 | |
6X2C
 
 | |
1DOX
 
 | | 1H AND 15N SEQUENTIAL ASSIGNMENT, SECONDARY STRUCTURE AND TERTIARY FOLD OF [2FE-2S] FERREDOXIN FROM SYNECHOCYSTIS SP. PCC 6803 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN [2FE-2S] | | Authors: | Lelong, C, Setif, P, Bottin, H, Andre, F, Neumann, J.M. | | Deposit date: | 1995-09-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N NMR sequential assignment, secondary structure, and tertiary fold of [2Fe-2S] ferredoxin from Synechocystis sp. PCC 6803.
Biochemistry, 34, 1995
|
|
1DOY
 
 | | 1H AND 15N SEQUENTIAL ASSIGNMENT, SECONDARY STRUCTURE AND TERTIARY FOLD OF [2FE-2S] FERREDOXIN FROM SYNECHOCYSTIS SP. PCC 6803 | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN [2FE-2S] | | Authors: | Lelong, C, Setif, P, Bottin, H, Andre, F, Neumann, J.M. | | Deposit date: | 1995-09-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N NMR sequential assignment, secondary structure, and tertiary fold of [2Fe-2S] ferredoxin from Synechocystis sp. PCC 6803.
Biochemistry, 34, 1995
|
|
3CV7
 
 | | Crystal structure of porcine aldehyde reductase ternary complex | | Descriptor: | 3,5-dichloro-2-hydroxybenzoic acid, Alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2008-04-18 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Structure of aldehyde reductase in ternary complex with coenzyme and the potent 20alpha-hydroxysteroid dehydrogenase inhibitor 3,5-dichlorosalicylic acid: Implications for inhibitor binding and selectivity
Arch.Biochem.Biophys., 479, 2008
|
|
6HTE
 
 | | Sulfolobus solfataricus Tryptophan Synthase B2a | | Descriptor: | DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE, Tryptophan synthase beta chain 2 | | Authors: | Fleming, J, Mayans, O, Bucher, R. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Evolutionary Morphing of Tryptophan Synthase: Functional Mechanisms for the Enzymatic Channeling of Indole.
J.Mol.Biol., 430, 2018
|
|
3DCI
 
 | | The Structure of a putative arylesterase from Agrobacterium tumefaciens str. C58 | | Descriptor: | ACETIC ACID, Arylesterase, CHLORIDE ION, ... | | Authors: | Cuff, M.E, Xu, X, Zheng, H, Binkowski, T.A, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-03 | | Release date: | 2008-09-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of a putative arylesterase from Agrobacterium tumefaciens str. C58
TO BE PUBLISHED
|
|
