3C2E
 
 | |
4FE2
 
 | | X-Ray Structure of SAICAR Synthetase (PurC) from Streptococcus pneumoniae complexed with AIR, ADP, Asp and Mg2+ | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOTIDE, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wolf, N, Abad-Zapatero, C, Johnson, M.E, Fung, L.M.-W. | | Deposit date: | 2012-05-29 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structures of SAICAR synthetase (PurC) from Streptococcus pneumoniae with ADP, Mg(2+), AIR and Asp.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4EW2
 
 | | The structure of human glycinamide ribonucleotide transformylase in complex with 10S-methylthio-DDATHF. | | Descriptor: | N-({4-[(1S)-4-(2,4-diamino-6-oxo-1,6-dihydropyrimidin-5-yl)-1-(methylsulfanyl)butyl]phenyl}carbonyl)-L-glutamic acid, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Connelly, S, DeMartino, K, Boger, D.L, Wilson, I.A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Biological and Structural Evaluation of 10R- and 10S-Methylthio-DDACTHF Reveals a New Role for Sulfur in Inhibition of Glycinamide Ribonucleotide Transformylase.
Biochemistry, 52, 2013
|
|
1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1IBJ
 
 | | Crystal structure of cystathionine beta-lyase from Arabidopsis thaliana | | Descriptor: | CARBONATE ION, CYSTATHIONINE BETA-LYASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Breitinger, U, Clausen, T, Messerschmidt, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of cystathionine beta-lyase from Arabidopsis and its substrate specificity
Plant Physiol., 126, 2001
|
|
2O4C
 
 | | Crystal Structure of D-Erythronate-4-phosphate Dehydrogenase Complexed with NAD | | Descriptor: | Erythronate-4-phosphate dehydrogenase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Ha, J.Y, Lee, J.H, Kim, K.H, Kim, D.J, Lee, H.H, Kim, H.K, Yoon, H.J, Suh, S.W. | | Deposit date: | 2006-12-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Erythronate-4-phosphate Dehydrogenase Complexed with NAD
J.Mol.Biol., 366, 2007
|
|
2NTM
 
 | |
2NTL
 
 | | Crystal structure of PurO/AICAR from Methanothermobacter thermoautotrophicus | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, IMP cyclohydrolase | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
2NTK
 
 | | Crystal structure of PurO/IMP from Methanothermobacter thermoautotrophicus | | Descriptor: | IMP cyclohydrolase, INOSINIC ACID | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
2QK4
 
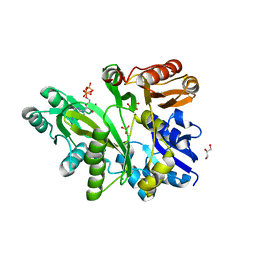 | | Human glycinamide ribonucleotide synthetase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lehtio, L, Welin, M, Arrowsmith, C.H, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Herman, M.D, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural studies of tri-functional human GART.
Nucleic Acids Res., 38, 2010
|
|
2IU0
 
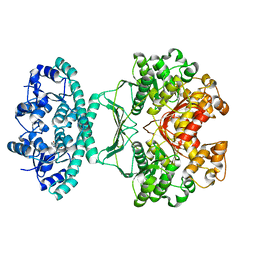 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, Onofrio, A.D, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase
J.Biol.Chem., 282, 2007
|
|
2IU3
 
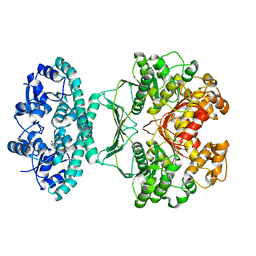 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase.
J. Biol. Chem., 282, 2007
|
|
3QW3
 
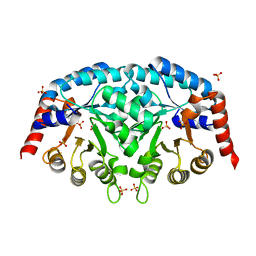 | | Structure of Leishmania donovani OMP decarboxylase | | Descriptor: | Orotidine-5-phosphate decarboxylase/orotate phosphoribosyltransferase, putative (Ompdcase-oprtase, putative), ... | | Authors: | French, J.B, Ealick, S.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization.
J.Biol.Chem., 286, 2011
|
|
2GQR
 
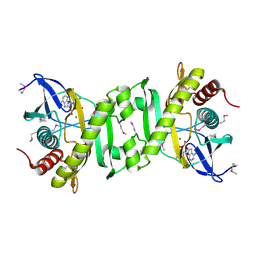 | | SAICAR Synthetase Complexed with ADP-Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Ginder, N.D, Honzatko, R.B. | | Deposit date: | 2006-04-21 | | Release date: | 2006-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide Complexes of Escherichia coli Phosphoribosylaminoimidazole Succinocarboxamide Synthetase.
J.Biol.Chem., 281, 2006
|
|
2GQS
 
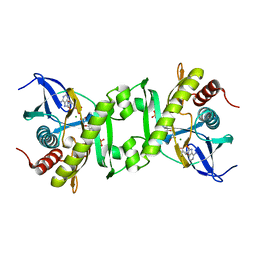 | | SAICAR Synthetase Complexed with CAIR-Mg2+ and ADP | | Descriptor: | 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, ADENOSINE-5'-DIPHOSPHATE, FORMIC ACID, ... | | Authors: | Ginder, N.D, Honzatko, R.B. | | Deposit date: | 2006-04-21 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Nucleotide Complexes of Escherichia coli Phosphoribosylaminoimidazole Succinocarboxamide Synthetase.
J.Biol.Chem., 281, 2006
|
|
3E9K
 
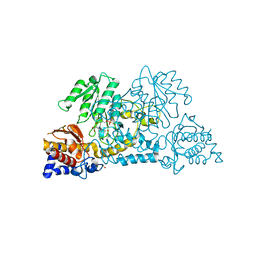 | | Crystal structure of Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex | | Descriptor: | 3-Hydroxyhippuric acid, Kynureninase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lima, S, Kumar, S, Gawandi, V, Momany, C, Phillips, R.S. | | Deposit date: | 2008-08-22 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex: insights into the molecular basis of kynureninase substrate specificity.
J.Med.Chem., 52, 2009
|
|
3ETJ
 
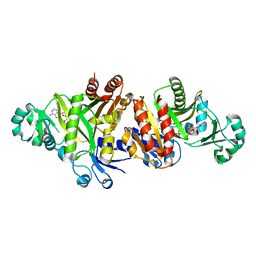 | | Crystal structure E. coli Purk in complex with Mg, ADP, and Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, HYDROGENPHOSPHATE ION, ... | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2008-10-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of the active site geometry of N(5)-Carboxyaminoimidazole ribonucleotide synthetase from Escherichia coli.
Biochemistry, 47, 2008
|
|
3DEZ
 
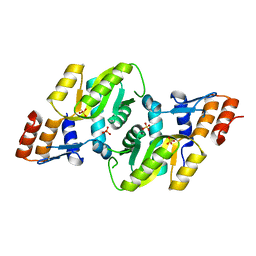 | | Crystal structure of Orotate phosphoribosyltransferase from Streptococcus mutans | | Descriptor: | Orotate phosphoribosyltransferase, SULFATE ION | | Authors: | Liu, C.P, Gao, Z.Q, Hou, H.F, Li, L.F, Su, X.D, Dong, Y.H. | | Deposit date: | 2008-06-11 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of orotate phosphoribosyltransferase from the caries pathogen Streptococcus mutans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3ETH
 
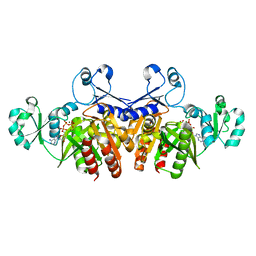 | | Crystal structure of E. coli Purk in complex with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Phosphoribosylaminoimidazole carboxylase ATPase subunit | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2008-10-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of the active site geometry of N(5)-Carboxyaminoimidazole ribonucleotide synthetase from Escherichia coli.
Biochemistry, 47, 2008
|
|
6XSS
 
 | | CryoEM structure of designed helical fusion protein C4_nat_HFuse-7900 | | Descriptor: | C4_nat_HFuse-7900 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
6XT4
 
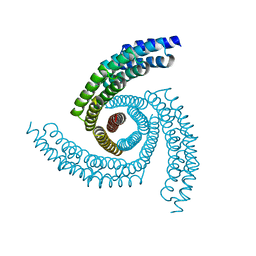 | |
6XNS
 
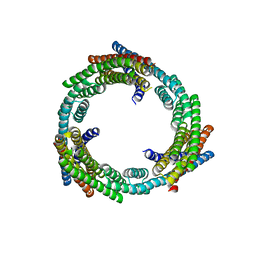 | | C3_crown-05 | | Descriptor: | C3_crown-05 | | Authors: | Bick, M.J, Hsia, Y, Sankaran, B, Baker, D. | | Deposit date: | 2020-07-04 | | Release date: | 2020-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
5IEN
 
 | | Structure of CDL2.2, a computationally designed Vitamin-D3 binder | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.2, GLYCEROL | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
5IEO
 
 | | Structure of CDL2.3a, a computationally designed Vitamin-D3 binder | | Descriptor: | 1,2-ETHANEDIOL, 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.3a | | Authors: | Stoddard, B.L, Doyle, L.A. | | Deposit date: | 2016-02-25 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
2RT4
 
 | |
