6TTA
 
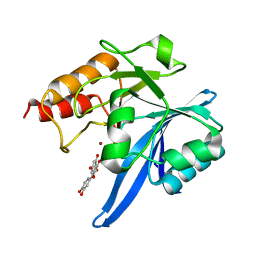 | | Haddock model of NDM-1/quercetin complex | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
3CVL
 
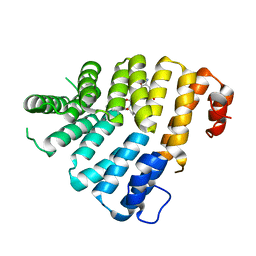 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphofructokinase (PFK) PTS1 peptide | | Descriptor: | Peroxisome targeting signal 1 receptor PEX5, T. brucei PFK PTS1 peptide Ac-HEELAKL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
5DFI
 
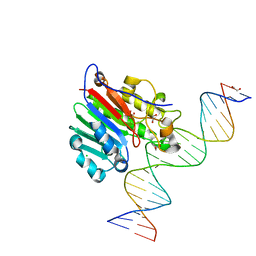 | | Human APE1 phosphorothioate substrate complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(OMC)P*(48Z)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3CXP
 
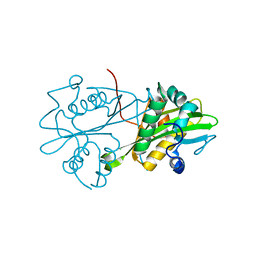 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 mutant E156A | | Descriptor: | CHLORIDE ION, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
6KMP
 
 | | 100K X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, Protease | | Authors: | Das, A, Kovalevsky, A. | | Deposit date: | 2019-07-31 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
4MOT
 
 | | Structure of Streptococcus pneumonia pare in complex with AZ13072886 | | Descriptor: | 1-[4-(3-methylbutyl)-5-oxo-6-(pyridin-3-yl)-4,5-dihydro[1,3]thiazolo[5,4-b]pyridin-2-yl]-3-prop-2-en-1-ylurea, Topoisomerase IV subunit B | | Authors: | Ogg, D, Boriack-Sjodin, P.A. | | Deposit date: | 2013-09-12 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thiazolopyridone ureas as DNA gyrase B inhibitors: Optimization of antitubercular activity and efficacy.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3KQ0
 
 | | Crystal structure of human alpha1-acid glycoprotein | | Descriptor: | (2R)-2,3-dihydroxypropyl acetate, Alpha-1-acid glycoprotein 1, CHLORIDE ION | | Authors: | Schiefner, A, Schonfeld, D.L, Ravelli, R.B.G, Mueller, U, Skerra, A. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8-A crystal structure of alpha1-acid glycoprotein (Orosomucoid) solved by UV RIP reveals the broad drug-binding activity of this human plasma lipocalin.
J.Mol.Biol., 384, 2008
|
|
7TCN
 
 | |
4DGM
 
 | | Crystal Structure of maize CK2 in complex with the inhibitor apigenin | | Descriptor: | 1,2-ETHANEDIOL, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Casein kinase II subunit alpha, ... | | Authors: | Lolli, G, Mazzorana, M, Battistutta, R. | | Deposit date: | 2012-01-26 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of protein kinase CK2 by flavonoids and tyrphostins. A structural insight.
Biochemistry, 51, 2012
|
|
6A3T
 
 | | The crystal structure of Mandelate oxidase R163L with 2-hydroxy-phenylacetamide | | Descriptor: | 1-{5-[(1S)-2-amino-1-hydroxy-2-oxo-1-phenylethyl]-7,8-dimethyl-2,4-dioxo-1,2,3,4-tetrahydrobenzo[g]pteridine-5,10-diium-10-yl}-1-deoxy-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-16 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.511 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4ZND
 
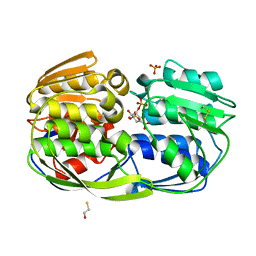 | | 2.55 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate, phosphate, and potassium | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, BETA-MERCAPTOETHANOL, PHOSPHATE ION, ... | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 2.55 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate, phosphate, and potassium
To Be Published
|
|
6TOD
 
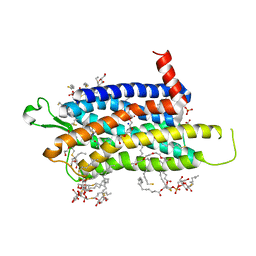 | | Crystal structure of the Orexin-1 receptor in complex with EMPA | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TQ9
 
 | | Crystal structure of the Orexin-1 receptor in complex with SB-408124 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1-[6,8-bis(fluoranyl)-2-methyl-quinolin-4-yl]-3-[4-(dimethylamino)phenyl]urea, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
5TTU
 
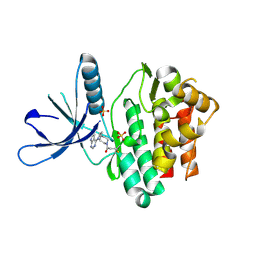 | | Jak3 with covalent inhibitor 7 | | Descriptor: | 1-[(3aR,7aR)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)octahydro-6H-pyrrolo[2,3-c]pyridin-6-yl]propan-1-one, SULFATE ION, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans.
J. Med. Chem., 60, 2017
|
|
6TOT
 
 | | Crystal structure of the Orexin-1 receptor in complex with lemborexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (1~{R},2~{S})-2-[(2,4-dimethylpyrimidin-5-yl)oxymethyl]-~{N}-(5-fluoranylpyridin-2-yl)-2-(3-fluorophenyl)cyclopropane-1-carboxamide, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
3FVF
 
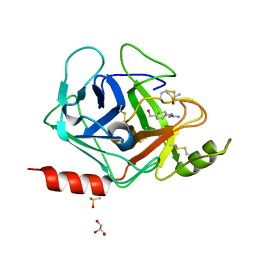 | | The Crystal Structure of Prostasin Complexed with Camostat at 1.6 Angstroms Resolution | | Descriptor: | 1-[4-(hydroxymethyl)phenyl]guanidine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Spraggon, G, Hornsby, M, Shipway, A, Harris, J.L, Lesley, S.A. | | Deposit date: | 2009-01-15 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations.
Protein Sci., 18, 2009
|
|
4ZK4
 
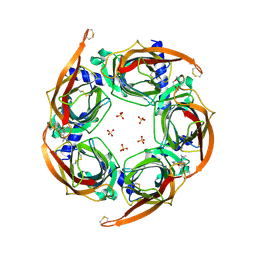 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with 7-(5-isopropoxy-pyridin-3-yl)-1-methyl-1,7-diaza-spiro[4.4]nonane | | Descriptor: | (5R)-1-methyl-7-[5-(propan-2-yloxy)pyridin-3-yl]-1,7-diazaspiro[4.4]nonane, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bobango, J, Wu, J, Talley, T.T. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with 7-(5-isopropoxy-pyridin-3-yl)-1-methyl-1,7-diaza-spiro[4.4]nonane.
To be Published
|
|
2R83
 
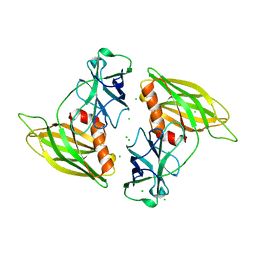 | | Crystal structure analysis of human synaptotagmin 1 C2A-C2B | | Descriptor: | CHLORIDE ION, Synaptotagmin-1 | | Authors: | Sutton, R.B, Fuson, K.L, Montes, M, Robert, J.J. | | Deposit date: | 2007-09-10 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of human synaptotagmin 1 C2AB in the absence of Ca2+ reveals a novel domain association.
Biochemistry, 46, 2007
|
|
9BTI
 
 | | Rhesus Fab 40591-a.01 in complex with T250.4 RnS SOSIP Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-05-15 | | Release date: | 2025-06-11 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural and genetic basis of HIV-1 envelope V2 apex recognition by rhesus broadly neutralizing antibodies.
J.Exp.Med., 222, 2025
|
|
7X87
 
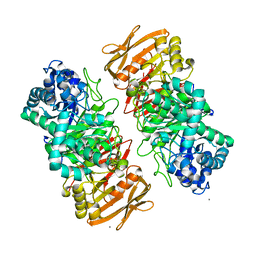 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with sophotetraose observed as sophorose | | Descriptor: | Beta-galactosidase, CALCIUM ION, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J Biol Chem, 298, 2022
|
|
7X78
 
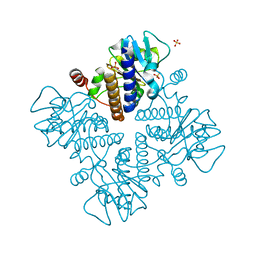 | | L-fuculose 1-phosphate aldolase | | Descriptor: | L-fuculose phosphate aldolase, MAGNESIUM ION, SULFATE ION | | Authors: | Lou, X, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of an L-fuculose-1-phosphate aldolase from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 607, 2022
|
|
6TT8
 
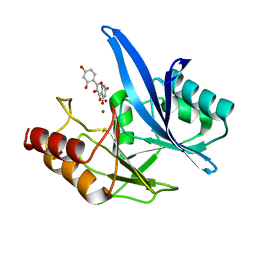 | | Haddock model of NDM-1/morin complex | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
3OP5
 
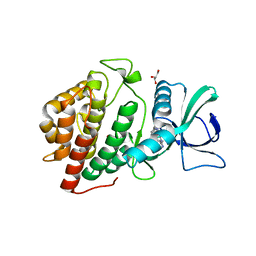 | | Human vaccinia-related kinase 1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase VRK1, ... | | Authors: | Allerston, C.K, Uttarkar, S, Savitsky, P, Elkins, J.M, Filippakopoulos, P, Krojer, T, Rellos, P, Fedorov, O, Eswaran, J, Brenner, B, Keates, T, Das, S, King, O, Chalk, R, Berridge, G, von Delft, F, Gileadi, O, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of human Vaccinia-Related Kinases (VRK) bound to small-molecule inhibitors identifies different P-loop conformations.
Sci Rep, 7, 2017
|
|
6TP3
 
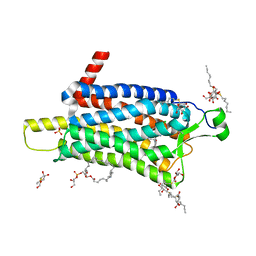 | | Crystal structure of the Orexin-1 receptor in complex with daridorexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Orexin receptor type 1, SULFATE ION, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
3CIV
 
 | |
