1NR8
 
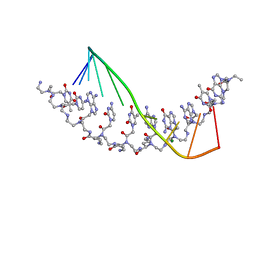 | | The crystal structure of a D-Lysine-based chiral PNA-DNA duplex | | Descriptor: | 5'-D(P*AP*GP*TP*GP*AP*TP*CP*TP*AP*C)-3', H-((GPN)*(TPN)*(APN)*(GPN)*(A66)*(T66)*(C66)*(APN)*(CPN)*(TPN))-NH2, MAGNESIUM ION | | Authors: | Menchise, V, De Simone, G, Tedeschi, T, Corradini, R, Sforza, S, Marchelli, R, Capasso, D, Saviano, M, Pedone, C. | | Deposit date: | 2003-01-24 | | Release date: | 2003-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights into peptide nucleic acid (PNA) structural features: The
crystal structure of a D-lysine-based chiral PNA-DNA duplex
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5T68
 
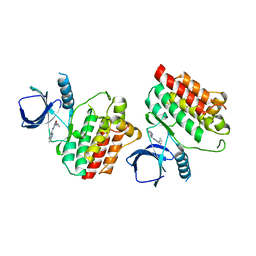 | | Crystal structure of Syk catalytic domain in complex with a furo[3,2-d]pyrimidine | | Descriptor: | N~4~-cyclopropyl-N~2~-(3-methyl-1H-indazol-6-yl)furo[3,2-d]pyrimidine-2,4-diamine, Tyrosine-protein kinase SYK | | Authors: | Argiriadi, M.A, Hoemann, M, Wilson, N, Banach, D, Burchat, A, Calderwood, D, Clapham, B, Cox, P, Duignan, D.B, Konopacki, D, Somal, G, Vasudevan, A. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Synthesis and optimization of furano[3,2-d]pyrimidines as selective spleen tyrosine kinase (Syk) inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
2BIU
 
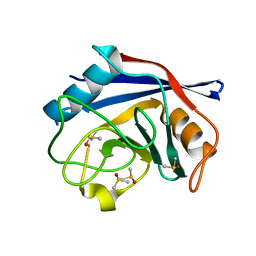 | | Crystal structure of human cyclophilin D at 1.7 A resolution, DMSO complex | | Descriptor: | DIMETHYL SULFOXIDE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Hennig, M, Thoma, R, Stihle, M, Schlatter, D. | | Deposit date: | 2005-01-26 | | Release date: | 2005-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Engineering Yields Crystals of Cyclophilin D Diffracting to 1.7 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4G4J
 
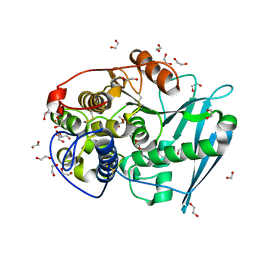 | | Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile in complex with methyl 4-O-methyl-beta-D-glucopyranuronate determined at 2.35 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-glucuronoyl methylesterase, GLYCEROL, ... | | Authors: | Charavgi, M.D, Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2012-07-16 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4AZC
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | (2S,3aR,5R,6S,7R,7aR)-5-(hydroxymethyl)-2-methyl-2,3a,5,6,7,7a-hexahydro-1H-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
4GSU
 
 | | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Probable conserved lipoprotein LPPS | | Authors: | Kim, H.S, Kim, J, Im, H.N, Yoon, J.Y, An, D.R, Yoon, H.J, Kim, J.Y, Min, H.K, Kim, S.-J, Lee, J.Y, Han, B.W, Suh, S.W. | | Deposit date: | 2012-08-28 | | Release date: | 2013-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GSR
 
 | | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains | | Descriptor: | 2-(ETHYLMERCURI-THIO)-BENZOIC ACID, GLYCEROL, Probable conserved lipoprotein LPPS | | Authors: | Kim, H.S, Kim, J, Im, H.N, Yoon, J.Y, An, D.R, Yoon, H.J, Kim, J.Y, Min, H.K, Kim, S.-J, Lee, J.Y, Han, B.W, Suh, S.W. | | Deposit date: | 2012-08-28 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis for the inhibition of Mycobacterium tuberculosis L,D-transpeptidase by meropenem, a drug effective against extensively drug-resistant strains
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3KCO
 
 | |
3KBN
 
 | |
2XV7
 
 | | Crystal structure of vascular endothelial growth factor D | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR D, alpha-D-mannopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Leppanen, V.-M, Jeltsch, M, Anisimov, A, Tvorogov, D, Aho, K, Kalkkinen, N, Toivanen, P, Yla-Herttuala, S, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2010-10-23 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Determinants of Vascular Endothelial Growth Factor-D - Receptor Binding and Specificity.
Blood, 117, 2011
|
|
7LL8
 
 | | D-Protein RFX-V1 Bound to the VEGFR1 Domain 2 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V1 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
7LL9
 
 | | D-Protein RFX-V2 Bound to the VEGFR1 Domain 3 Site on VEGF-A | | Descriptor: | Isoform L-VEGF189 of Vascular endothelial growth factor A, RFX-V2 | | Authors: | Marinec, P.S, Landgraf, K.E, Uppalapati, M, Chen, G, Xie, D, Jiang, Q, Zhao, Y, Petriello, A, Deshayes, K, Kent, S.B.H, Ault-Riche, D, Sidhu, S.S. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Non-immunogenic Bivalent d-Protein Potently Inhibits Retinal Vascularization and Tumor Growth.
Acs Chem.Biol., 16, 2021
|
|
3WGZ
 
 | | Crystal structure of meso-dapdh Q154L/T173I/R199M/P248S/H249N/N276S mutant with D-leucine of from Clostridium tetani E88 | | Descriptor: | D-LEUCINE, Meso-diaminopimelate dehydrogenase | | Authors: | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2013-08-20 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of meso-dapdh Q154L/T173I/R199M/P248S/H249N/N276S mutant with D-leucine of from Clostridium tetani E88
to be published
|
|
3BD6
 
 | | Glycogen Phosphorylase complex with 1(-D-ribofuranosyl) cyanuric acid | | Descriptor: | 1-beta-D-ribofuranosyl-1,3,5-triazinane-2,4,6-trione, Glycogen phosphorylase, muscle form | | Authors: | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
4P7R
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with a poly-beta-1,6-N-acetyl-D-glucosamine (PNAG) hexamer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1L2G
 
 | | Structure of a C-terminally truncated form of glycoprotein D from HSV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein D | | Authors: | Carfi, A, Willis, S.H, Whitbeck, J.C, Krummenacher, C, Cohen, G.H, Eisenberg, R.J, Wiley, D.C. | | Deposit date: | 2002-02-21 | | Release date: | 2003-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Herpes simplex virus glycoprotein D bound to the human receptor HveA.
Mol.Cell, 8, 2001
|
|
4AVS
 
 | |
1XIE
 
 | |
4OAK
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Alanine-D-Alanine and copper (II) | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1XLG
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | ALUMINUM ION, D-XYLOSE ISOMERASE, MAGNESIUM ION, ... | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
2XWB
 
 | | Crystal Structure of Complement C3b in complex with Factors B and D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3B ALPHA' CHAIN, ... | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
4CSC
 
 | |
1F0X
 
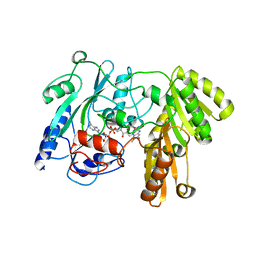 | | CRYSTAL STRUCTURE OF D-LACTATE DEHYDROGENASE, A PERIPHERAL MEMBRANE RESPIRATORY ENZYME. | | Descriptor: | D-LACTATE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Dym, O, Pratt, E.A, Ho, C, Eisenberg, D. | | Deposit date: | 2000-05-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KNJ
 
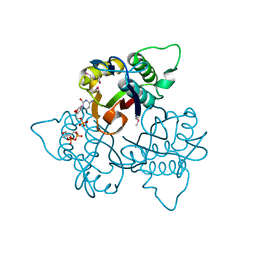 | | Co-Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli Involved in Mevalonate-Independent Isoprenoid Biosynthesis, Complexed with CMP/MECDP/Mn2+ | | Descriptor: | 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE, 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
1UHV
 
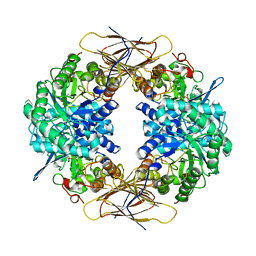 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-11 | | Release date: | 2003-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
