5Z21
 
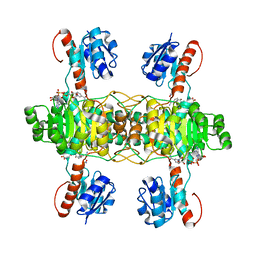 | | The ternary structure of D-lactate dehydrogenase from Fusobacterium nucleatum with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-lactate dehydrogenase, OXAMIC ACID | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
3K5P
 
 | |
5Z1Z
 
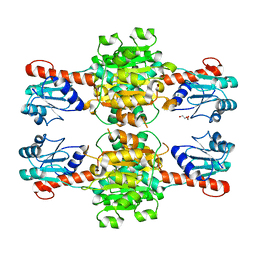 | | The apo-structure of D-lactate dehydrogenase from Escherichia coli | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.
Biochemistry, 57, 2018
|
|
7JP2
 
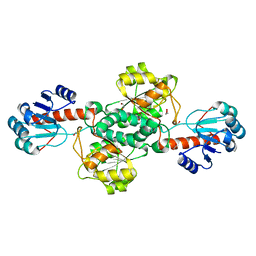 | | Crystal structure of TP0037 from Treponema pallidum, a D-lactate dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-lactate dehydrogenase | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Biophysical and Biochemical Characterization of TP0037, a d-Lactate Dehydrogenase, Supports an Acetogenic Energy Conservation Pathway in Treponema pallidum.
Mbio, 11, 2020
|
|
8IQ7
 
 | |
4NU6
 
 | | Crystal Structure of PTDH R301K | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Phosphonate dehydrogenase, SULFATE ION | | Authors: | Nair, S.K, Chekan, J.R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Chemical rescue and inhibition studies to determine the role of arg301 in phosphite dehydrogenase.
Plos One, 9, 2014
|
|
8I5Z
 
 | |
3N7U
 
 | | NAD-dependent formate dehydrogenase from higher-plant Arabidopsis thaliana in complex with NAD and azide | | Descriptor: | AZIDE ION, Formate dehydrogenase, GLYCEROL, ... | | Authors: | Shabalin, I.G, Polyakov, K.M, Serov, A.E, Skirgello, O.E, Sadykhov, E.G, Dorovatovskiy, P.V, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the apo and holo forms of NAD-dependent formate dehydrogenase from the higher-plant Arabidopsis Thaliana
To be Published
|
|
6ABI
 
 | | The apo-structure of D-lactate dehydrogenase from Fusobacterium nucleatum | | Descriptor: | D-lactate dehydrogenase, GLYCEROL, SULFATE ION | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
6ABJ
 
 | | The apo-structure of D-lactate dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, D-lactate dehydrogenase (Fermentative) | | Authors: | Furukawa, N, Miyanaga, A, Nakajima, M, Taguchi, H. | | Deposit date: | 2018-07-21 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria
Biochemistry, 57, 2018
|
|
6BII
 
 | | Crystal Structure of Pyrococcus yayanosii Glyoxylate Hydroxypyruvate Reductase in complex with NADP and malonate (re-refinement of 5AOW) | | Descriptor: | GLYCEROL, Glyoxylate reductase, MALONATE ION, ... | | Authors: | Lassalle, L, Shabalin, I.G, Girard, E, Minor, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights into the mechanism of substrates trafficking in Glyoxylate/Hydroxypyruvate reductases.
Sci Rep, 6, 2016
|
|
7KWM
 
 | | CtBP1 (28-375) L182F/V185T - AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, C-terminal-binding protein 1, CALCIUM ION | | Authors: | Royer, W.E, Del Campo, M. | | Deposit date: | 2020-12-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NAD(H) phosphates mediate tetramer assembly of human C-terminal binding protein (CtBP).
J.Biol.Chem., 296, 2021
|
|
3NAQ
 
 | | Apo-form of NAD-dependent formate dehydrogenase from higher-plant Arabidopsis thaliana | | Descriptor: | Formate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Polyakov, K.M, Serov, A.E, Skirgello, O.E, Sadykhov, E.G, Dorovatovskiy, P.V, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the apo and holo forms of NAD-dependent formate dehydrogenase from the higher-plant Arabidopsis Thaliana
to be published
|
|
3EVT
 
 | | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum | | Descriptor: | Phosphoglycerate dehydrogenase | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of phosphoglycerate dehydrogenase from Lactobacillus plantarum
To be Published
|
|
4EBF
 
 | |
4E5K
 
 | |
6IH8
 
 | |
2CUK
 
 | |
6PEX
 
 | |
6PLF
 
 | | Crystal structure of human PHGDH complexed with Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-{(1S)-1-[(5-chloro-6-{[(5S)-2-oxo-1,3-oxazolidin-5-yl]methoxy}-1H-indole-2-carbonyl)amino]-2-hydroxyethyl}benzoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Olland, A, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2019-06-30 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of 3-phosphoglycerate dehydrogenase (PHGDH) by indole amides abrogates de novo serine synthesis in cancer cells.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
2D0I
 
 | |
6JX1
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101 | | Descriptor: | Formate dehydrogenase, GLYCEROL | | Authors: | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JUK
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | Deposit date: | 2019-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JUJ
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | Deposit date: | 2019-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
4DGS
 
 | | The crystals structure of dehydrogenase from Rhizobium meliloti | | Descriptor: | Dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-26 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystals structure of dehydrogenase from Rhizobium meliloti
To be Published
|
|
