3SUI
 
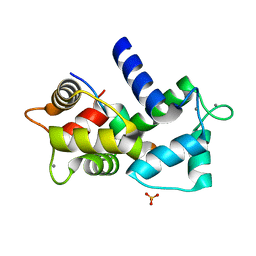 | |
3SX2
 
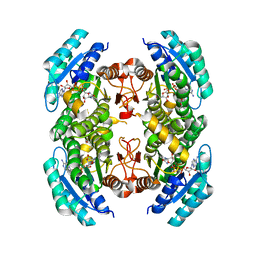 | |
3T6G
 
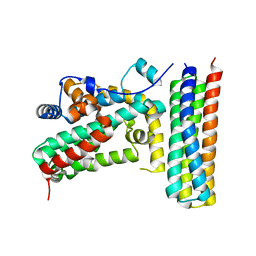 | |
3T2B
 
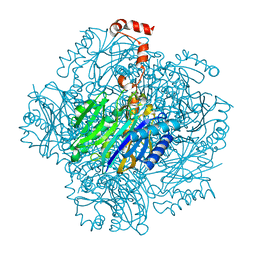 | | Fructose-1,6-bisphosphate aldolase/phosphatase from Thermoproteus neutrophilus, ligand free | | Descriptor: | Fructose-1,6-bisphosphate aldolase/phosphatase, MAGNESIUM ION | | Authors: | Du, J, Say, R, Lue, W, Fuchs, G, Einsle, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
3SOH
 
 | | Architecture of the Flagellar Rotor | | Descriptor: | Flagellar motor switch protein FliG, Flagellar motor switch protein FliM | | Authors: | Koushik, P, Gonzalez-Bonet, G, Bilwes, A.M, Crane, B.R, Blair, D. | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the flagellar rotor.
Embo J., 30, 2011
|
|
3T1I
 
 | | Crystal Structure of Human Mre11: Understanding Tumorigenic Mutations | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Double-strand break repair protein MRE11A, GLYCEROL, ... | | Authors: | Park, Y.B, Chae, J, Kim, Y, Cho, Y. | | Deposit date: | 2011-07-22 | | Release date: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human mre11: understanding tumorigenic mutations
Structure, 19, 2011
|
|
3VC0
 
 | | Crystal structure of Taipoxin beta subunit isoform 1 | | Descriptor: | Phospholipase A2 homolog, taipoxin beta chain | | Authors: | Cendron, L, Micetic, I, Polverino de Laureto, P, Beltramini, M, Paoli, M. | | Deposit date: | 2012-01-03 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of trimeric phospholipase A(2) neurotoxin from the Australian taipan snake venom.
Febs J., 279, 2012
|
|
3UT5
 
 | | Tubulin-Colchicine-Ustiloxin: Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ranaivoson, F.M, Gigant, B, Knossow, M. | | Deposit date: | 2011-11-25 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural plasticity of tubulin assembly probed by vinca-domain ligands.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3V6B
 
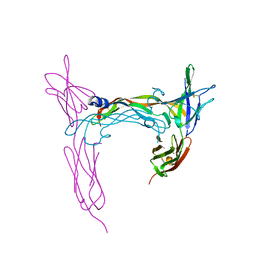 | | VEGFR-2/VEGF-E complex structure | | Descriptor: | VEGF-E, Vascular endothelial growth factor receptor 2 | | Authors: | Brozzo, M.S, Leppanen, V.-M, Winkler, F.K, Kisko, K, Ballmer-Hofer, K. | | Deposit date: | 2011-12-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Thermodynamic and structural description of allosterically regulated VEGFR-2 dimerization.
Blood, 119, 2012
|
|
3V4X
 
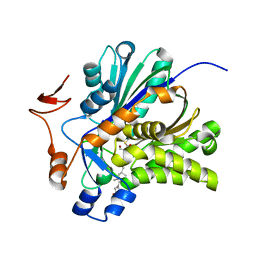 | | The Biochemical and Structural Basis for Inhibition of Enterococcus faecalis HMG-CoA Synthase, mvaS, by Hymeglusin | | Descriptor: | (7R,12R,13R)-13-formyl-12,14-dihydroxy-3,5,7-trimethyltetradeca-2,4-dienoic acid, HMG-CoA synthase | | Authors: | Skaff, D.A, Ramyar, K.X, McWhorter, W.J, Geisbrecht, B.V, Miziorko, H.M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Biochemical and structural basis for inhibition of Enterococcus faecalis hydroxymethylglutaryl-CoA synthase, mvaS, by hymeglusin.
Biochemistry, 51, 2012
|
|
3V64
 
 | | Crystal Structure of agrin and LRP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low-density lipoprotein receptor-related protein 4, ... | | Authors: | Zong, Y, Zhang, B, Gu, S, Lee, K, Zhou, J, Yao, G, Figueiedo, D, Perry, K, Mei, L, Jin, R. | | Deposit date: | 2011-12-18 | | Release date: | 2012-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of agrin-LRP4-MuSK signaling.
Genes Dev., 26, 2012
|
|
3VHN
 
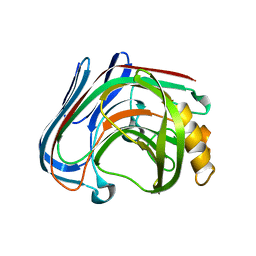 | | Y61G mutant of Cellulase 12A from thermotoga maritima | | Descriptor: | Endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Cheng, Y.-S, Ko, T.-P, Guo, R.-T, Liu, J.-R. | | Deposit date: | 2011-08-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enhanced activity of Thermotoga maritima cellulase 12A by mutating a unique surface loop
Appl.Microbiol.Biotechnol., 95, 2012
|
|
3VCP
 
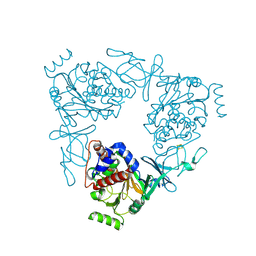 | | The 2.2 Angstrom structure of Stc2 with proline bound in the active site | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, PROLINE, ... | | Authors: | Daughtry, K.D, Xiao, Y, Stoner-Ma, D, Cho, E, Orville, A.M, Liu, P, Allen, K.N. | | Deposit date: | 2012-01-04 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman, and UV-Visible Spectroscopic Analysis of a Rieske-Type Demethylase.
J.Am.Chem.Soc., 134, 2012
|
|
3UXU
 
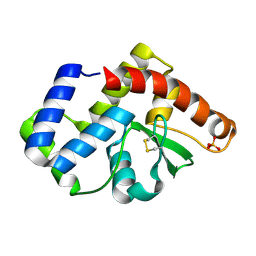 | |
3V4Y
 
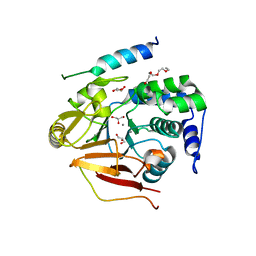 | | Crystal Structure of the first Nuclear PP1 holoenzyme | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Nuclear inhibitor of protein phosphatase 1, ... | | Authors: | Page, R, Peti, W, O'Connell, N.E, Nichols, S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | The Molecular Basis for Substrate Specificity of the Nuclear NIPP1:PP1 Holoenzyme.
Structure, 20, 2012
|
|
3UYB
 
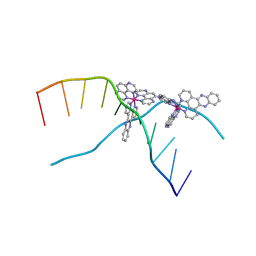 | | X-ray crystal structure of the ruthenium complex [Ru(tap)2(dppz)]2+ bound to d(TCGGTACCGA) | | Descriptor: | 5'-D(*TP*CP*GP*GP*TP*AP*CP*CP*GP*A)-3', BARIUM ION, Ru(tap)2(dppz) complex | | Authors: | Niyazi, H.N, Texeira, S.C.M, Mitchell, E.P, Cardin, J.C, Forsyth, V.T. | | Deposit date: | 2011-12-06 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of lambda-[Ru(TAP)2(dppz)]2+ with d(TCGGTACCGA)2 displaying kinking of DNA via semiintercalation.
To be Published
|
|
3USB
 
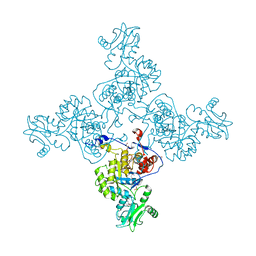 | | Crystal Structure of Bacillus anthracis Inosine Monophosphate Dehydrogenase in the complex with IMP | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Zhang, R, Wu, R, Gu, M, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-07 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3U8P
 
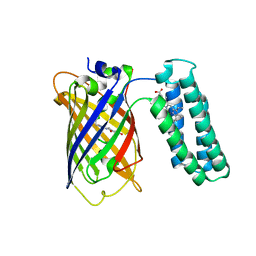 | | Cytochrome b562 integral fusion with EGFP | | Descriptor: | Cytochrome b562 integral fusion with enhanced green fluorescent protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arpino, J, Czapinska, H, Piasecka, A, Edwards, W.R, Barker, P, Gajda, M, Bochtler, M, Jones, D.D. | | Deposit date: | 2011-10-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for efficient chromophore communication and energy transfer in a constructed didomain protein scaffold.
J.Am.Chem.Soc., 134, 2012
|
|
3U83
 
 | | Crystal structure of nectin-1 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Poliovirus receptor-related protein 1 | | Authors: | Zhang, N, Yan, J, Lu, G, Guo, Z, Fan, Z, Wang, J, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-10-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion.
Nat Commun, 2, 2011
|
|
3VID
 
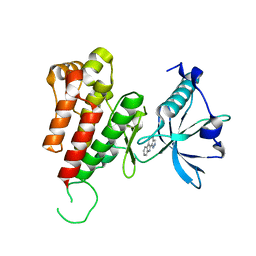 | | Crystal structure of human VEGFR2 kinase domain with Compound A. | | Descriptor: | 4,5,6,11-tetrahydro-1H-pyrazolo[4',3':6,7]cyclohepta[1,2-b]indole, Vascular endothelial growth factor receptor 2 | | Authors: | Iwata, H, Oki, H, Okada, K, Takagi, T, Tawada, M, Miyazaki, Y, Imamura, S, Hori, A, Hixon, M.S, Kimura, H, Miki, H. | | Deposit date: | 2011-09-29 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Back-to-Front Fragment-Based Drug Design Search Strategy Targeting the DFG-Out Pocket of Protein Tyrosine Kinases.
ACS MED.CHEM.LETT., 3, 2012
|
|
3VJM
 
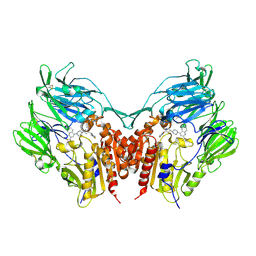 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #1 | | Descriptor: | 1,3-thiazolidin-3-yl[(2S,4S)-4-{4-[2-(trifluoromethyl)quinolin-4-yl]piperazin-1-yl}pyrrolidin-2-yl]methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fused bicyclic heteroarylpiperazine-substituted l-prolylthiazolidines as highly potent DPP-4 inhibitors lacking the electrophilic nitrile group
Bioorg.Med.Chem., 20, 2012
|
|
3US9
 
 | | Crystal Structure of the NCX1 Intracellular Tandem Calcium Binding Domains(CBD12) | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1 | | Authors: | Giladi, M, Sasson, Y, Hirsch, J.A, Khananshvili, D. | | Deposit date: | 2011-11-23 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | A common Ca2+-driven interdomain module governs eukaryotic NCX regulation.
Plos One, 7, 2012
|
|
3THX
 
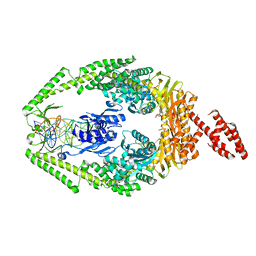 | | Human MutSbeta complexed with an IDL of 3 bases (Loop3) and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA Loop3 minus strand, DNA Loop3 plus strand, ... | | Authors: | Yang, W. | | Deposit date: | 2011-08-19 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops
Nat.Struct.Mol.Biol., 19, 2012
|
|
7LVQ
 
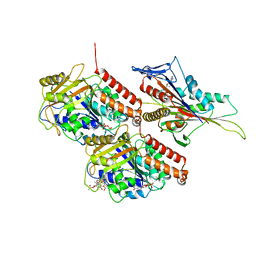 | | KIF14[391-743] - AMP-PNP closed state class in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF14, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Paydar, M, Dhakal, S, Kwok, B, Sosa, H. | | Deposit date: | 2021-02-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of mechano-chemical coupling by the mitotic kinesin KIF14.
Nat Commun, 12, 2021
|
|
7LVR
 
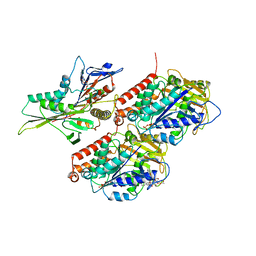 | | KIF14[391-743] - ADP-AlFx closed state class in complex with a microtubule | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Paydar, M, Dhakal, S, Kwok, B, Sosa, H. | | Deposit date: | 2021-02-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of mechano-chemical coupling by the mitotic kinesin KIF14.
Nat Commun, 12, 2021
|
|
