3G4F
 
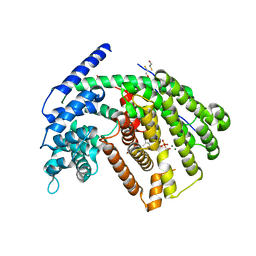 | | Crystal Structure of (+)- -Cadinene Synthase from Gossypium arboreum in complex with 2-fluorofarnesyl diphosphate | | Descriptor: | (+)-delta-cadinene synthase isozyme XC1, (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, BETA-MERCAPTOETHANOL, ... | | Authors: | Gennadios, H.A, Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2009-02-03 | | Release date: | 2009-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Crystal structure of (+)-delta-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis.
Biochemistry, 48, 2009
|
|
3EZO
 
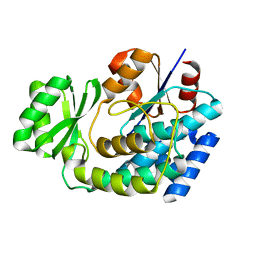 | |
3EY4
 
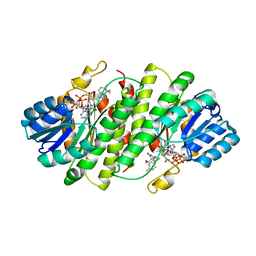 | | Further studies with the 2-amino-1,3-thiazol-4(5H)-one class of 11-hydroxysteroid dehydrogenase type 1 (11-HSD1) inhibitors: Reducing pregnane X receptor (PXR) activity and exploring activity in a monkey pharmacodynamic model | | Descriptor: | (5S)-2-{[(1S)-1-(4-fluorophenyl)ethyl]amino}-5-(1-hydroxy-1-methylethyl)-5-methyl-1,3-thiazol-4(5H)-one, 11-beta-Hydroxysteroid Dehydrogenase 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J.D, Jordan, S.R, Li, V. | | Deposit date: | 2008-10-17 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Further studies with the 2-amino-1,3-thiazol-4(5H)-one class of 11-hydroxysteroid dehydrogenase type 1 (11-HSD1) inhibitors: Reducing pregnane X receptor (PXR) activity and exploring activity in a monkey pharmacodynamic model
To be Published, 2008
|
|
3G50
 
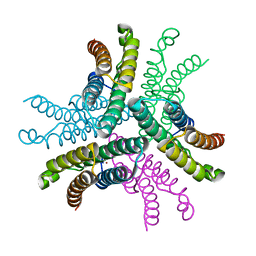 | | Crystal Structure of NiSOD D3A mutant at 1.9 A | | Descriptor: | NICKEL (II) ION, Superoxide dismutase [Ni] | | Authors: | Garman, S.C, Guce, A.I, Herbst, R.W, Bryngelson, P.A, Cabelli, D.E, Higgins, K.A, Ryan, K.C, Maroney, M.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of conserved tyrosine residues in NiSOD catalysis: a case of convergent evolution
Biochemistry, 48, 2009
|
|
3G7K
 
 | |
3G81
 
 | |
3FLR
 
 | |
3FPT
 
 | | The Crystal Structure of the Complex between Evasin-1 and CCL3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Evasin-1 | | Authors: | Shaw, J.P, Dias, J.M. | | Deposit date: | 2009-01-06 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of chemokine sequestration by a tick chemokine binding protein: the crystal structure of the complex between Evasin-1 and CCL3
Plos One, 4, 2009
|
|
3FQ4
 
 | |
3GMT
 
 | | Crystal structure of adenylate kinase from burkholderia pseudomallei | | Descriptor: | Adenylate kinase, SULFATE ION | | Authors: | Abendroth, J, Staker, B.L, Robinson, H, Buchko, G.W, Hewitt, S.N, Napuli, A.J, Van Voorhis, W, Stacy, R, Myler, P.J, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-03-15 | | Release date: | 2009-06-02 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of Burkholderia pseudomallei adenylate kinase (Adk): profound asymmetry in the crystal structure of the 'open' state.
Biochem.Biophys.Res.Commun., 394, 2010
|
|
3GT9
 
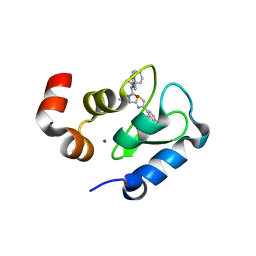 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | Baculoviral IAP repeat-containing 7, N-{(1S)-1-cyclohexyl-2-[(2S)-2-(4-naphthalen-1-yl-1,3-thiazol-2-yl)pyrrolidin-1-yl]-2-oxoethyl}-N~2~-methyl-L-alaninamide, ZINC ION | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antagonists of inhibitor of apoptosis proteins based on thiazole amide isosteres.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3GVF
 
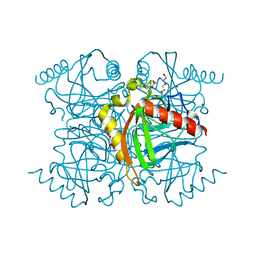 | |
3GW3
 
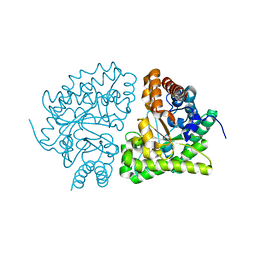 | | human UROD mutant K297N | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Hill, C.P, Phillips, J.D, Whitby, F.G, Warby, C, Kushner, J.P. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and kinetic characterization of mutant human uroporphyrinogen decarboxylases.
Cell Mol Biol (Noisy-le-grand), 55, 2009
|
|
3H5H
 
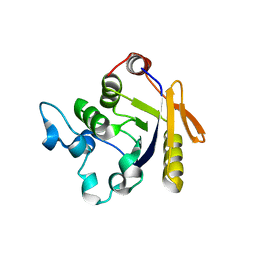 | |
3GW0
 
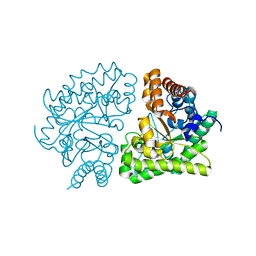 | | UROD mutant G318R | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Hill, C.P, Phillips, J.D, Whitby, F.G, Warby, C, Kushner, J.P. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of mutant human uroporphyrinogen decarboxylases.
Cell Mol Biol (Noisy-le-grand), 55, 2009
|
|
7YX9
 
 | |
7Z8E
 
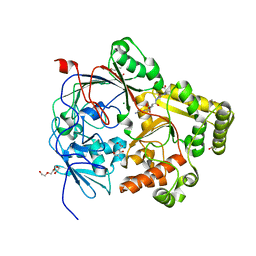 | | Crystal structure of the substrate-binding protein YejA from S. meliloti in complex with peptide fragment | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, GLY-SER-ASP-VAL-ALA, ... | | Authors: | Morera, S, Vigouroux, V, Travin, D.Y. | | Deposit date: | 2022-03-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual-Uptake Mode of the Antibiotic Phazolicin Prevents Resistance Acquisition by Gram-Negative Bacteria.
Mbio, 14, 2023
|
|
7YXB
 
 | | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, CLIP peptide, ... | | Authors: | Roske, Y, Abualrous, E.T, Freund, C. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange.
Nat.Chem.Biol., 19, 2023
|
|
7Z0Q
 
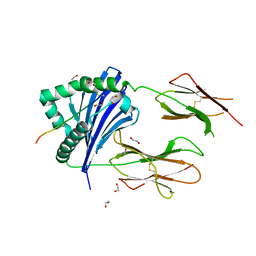 | | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange | | Descriptor: | 1,2-ETHANEDIOL, CLIP peptide, HLA class II histocompatibility antigen, ... | | Authors: | Roske, Y, Abualrous, E.T, Freund, C. | | Deposit date: | 2022-02-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange.
Nat.Chem.Biol., 19, 2023
|
|
7VUZ
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with PAMP-12, state2 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VV4
 
 | |
7VV5
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80, state1 | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VDH
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80, state2 | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
7VV0
 
 | |
7VV6
 
 | | Cryo-EM structure of pseudoallergen receptor MRGPRX2 complex with C48/80 (local) | | Descriptor: | 2-[4-methoxy-3-[[2-methoxy-3-[[2-methoxy-5-[2-(methylamino)ethyl]phenyl]methyl]-5-[2-(methylamino)ethyl]phenyl]methyl]phenyl]-~{N}-methyl-ethanamine, CHOLESTEROL, Mas-related G-protein coupled receptor member X2 | | Authors: | Li, Y, Yang, F. | | Deposit date: | 2021-11-04 | | Release date: | 2021-12-01 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure, function and pharmacology of human itch receptor complexes.
Nature, 600, 2021
|
|
