8P3O
 
 | |
8P3P
 
 | |
2LDL
 
 | | Solution NMR Structure of the HIV-1 Exon Splicing Silencer 3 | | Descriptor: | RNA (27-MER) | | Authors: | Mishler, C, Levengood, J.D, Johnson, C.A, Rajan, P, Znosko, B.M. | | Deposit date: | 2011-05-27 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the HIV-1 Exon Splicing Silencer 3.
J.Mol.Biol., 415, 2012
|
|
6NAJ
 
 | |
7DY7
 
 | | Discovery of Novel Small-molecule Inhibitors of PD-1/PD-L1 Axis that Promotes PD-L1 Internalization and Degradation | | Descriptor: | 2-[[3-[[5-(2-methyl-3-phenyl-phenyl)-1,3,4-oxadiazol-2-yl]amino]phenyl]methylamino]ethanol, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Wang, T.Y, Lu, M.L, Jiang, S, Xiao, Y.B. | | Deposit date: | 2021-01-20 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of Small-Molecule Inhibitors of the PD-1/PD-L1 Axis That Promote PD-L1 Internalization and Degradation.
J.Med.Chem., 65, 2022
|
|
3REZ
 
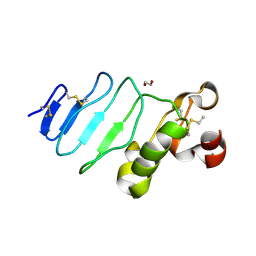 | | glycoprotein GPIb variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Platelet glycoprotein Ib beta chain, ... | | Authors: | McEwan, P.A, Yang, W, Carr, K.H, Mo, X, Zheng, X, Li, R, Emsley, J. | | Deposit date: | 2011-04-05 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the structures of GPIb beta and a GPIb beta/GPIX chimera
Blood, 118, 2011
|
|
3RFE
 
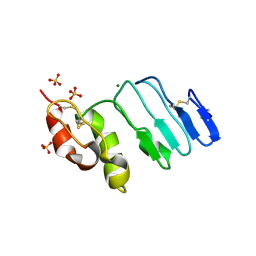 | | Crystal structure of glycoprotein GPIb ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Platelet glycoprotein Ib beta chain, ... | | Authors: | McEwan, P.A, Yang, W, Carr, K.H, Mo, X, Zheng, X, Li, R, Emsley, J. | | Deposit date: | 2011-04-06 | | Release date: | 2011-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.245 Å) | | Cite: | Quaternary organization of GPIb-IX complex and insights into Bernard-Soulier syndrome revealed by the structures of GPIbbeta and a GPIbbeta/GPIX chimer
Blood, 118, 2011
|
|
1HS4
 
 | |
4K27
 
 | |
2BN6
 
 | | P-Element Somatic Inhibitor Protein | | Descriptor: | PSI | | Authors: | Ignjatovic, T, Yang, J.C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
2BN5
 
 | | P-Element Somatic Inhibitor Protein Complex with U1-70k proline-rich peptide | | Descriptor: | PSI, U1 SMALL NUCLEAR RIBONUCLEOPROTEIN 70 KDA | | Authors: | Ignjatovic, T, Yang, J.-C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
6GG1
 
 | | Structure of PROSS-edited human interleukin 24 | | Descriptor: | Interleukin-24, NICKEL (II) ION, SULFATE ION | | Authors: | Kolenko, P, Zahradnik, J, Kolarova, L, Schneider, B. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Flexible regions govern promiscuous binding of IL-24 to receptors IL-20R1 and IL-22R1.
Febs J., 286, 2019
|
|
6KM7
 
 | | The structural basis for the internal interaction in RBBP5 | | Descriptor: | HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, Retinoblastoma-binding protein 5, ... | | Authors: | Han, J, Li, T, Chen, Y. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The internal interaction in RBBP5 regulates assembly and activity of MLL1 methyltransferase complex.
Nucleic Acids Res., 47, 2019
|
|
2MXS
 
 | | Solution NMR-structure of the neomycin sensing riboswitch RNA bound to paromomycin | | Descriptor: | PAROMOMYCIN, RNA (27-MER) | | Authors: | Schmidtke, S, Duchardt-Ferner, E, Ohlenschlaeger, O, Gottstein, D, Wohnert, J. | | Deposit date: | 2015-01-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5BT9
 
 | |
2ZXI
 
 | | Structure of Aquifex aeolicus GidA in the form II crystal | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2008-12-24 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved cysteine residues of GidA are essential for biogenesis of 5-carboxymethylaminomethyluridine at tRNA anticodon
Structure, 17, 2009
|
|
2ZXH
 
 | | Structure of Aquifex aeolicus GidA in the form I crystal | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2008-12-24 | | Release date: | 2009-05-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conserved cysteine residues of GidA are essential for biogenesis of 5-carboxymethylaminomethyluridine at tRNA anticodon
Structure, 17, 2009
|
|
4EUL
 
 | | Crystal structure of enhanced Green Fluorescent Protein to 1.35A resolution reveals alternative conformations for Glu222 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Green fluorescent protein, ... | | Authors: | Jones, D.D, Arpino, J.A.J, Rizkallah, P.J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of enhanced green fluorescent protein to 1.35 a resolution reveals alternative conformations for glu222.
Plos One, 7, 2012
|
|
5UCX
 
 | |
1XZP
 
 | | Structure of the GTP-binding protein TrmE from Thermotoga maritima | | Descriptor: | Probable tRNA modification GTPase trmE, SULFATE ION | | Authors: | Scrima, A, Vetter, I.R, Armengod, M.E, Wittinghofer, A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-01-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the TrmE GTP-binding protein and its implications for tRNA modification
Embo J., 24, 2005
|
|
1XZQ
 
 | | Structure of the GTP-binding protein TrmE from Thermotoga maritima complexed with 5-formyl-THF | | Descriptor: | N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, Probable tRNA modification GTPase trmE | | Authors: | Scrima, A, Vetter, I.R, Armengod, M.E, Wittinghofer, A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-01-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the TrmE GTP-binding protein and its implications for tRNA modification
Embo J., 24, 2005
|
|
7DN7
 
 | | Crystal structure of ternary complexes of lactoperoxidase with hydrogen peroxide at 1.70 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Singh, A.K, Singh, R.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a ternary complex of lactoperoxidase with iodide and hydrogen peroxide at 1.77 angstrom resolution.
J.Inorg.Biochem., 220, 2021
|
|
4LNS
 
 | |
3OK4
 
 | |
3B89
 
 | |
