8CRC
 
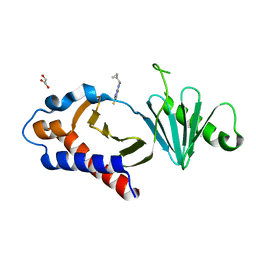 | | Structure of human Plk1 PBD in complex with Allopole-A | | Descriptor: | 7-chloro-4-(cyclopropylmethyl)-1-thioxo-2,4-dihydrothieno[2,3-e][1,2,4]triazolo[4,3-a]pyrimidin-5(1H)-one, GLYCEROL, Serine/threonine-protein kinase PLK1 | | Authors: | Kirsch, K, Park, J.E, Lee, K.S. | | Deposit date: | 2023-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Specific inhibition of an anticancer target, polo-like kinase 1, by allosterically dismantling its mechanism of substrate recognition.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FIZ
 
 | | Cryo-EM structure of E. coli 70S Ribosome containing mRNA and tRNA (in the transcription-translation complex) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
8F67
 
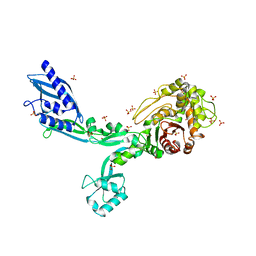 | | Crystal structure of the refolded Penicillin Binding Protein 5 (PBP5) of Enterococcus faecium | | Descriptor: | Pbp5, SULFATE ION | | Authors: | D'Andrea, E.D, Schoenle, M.V, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3I
 
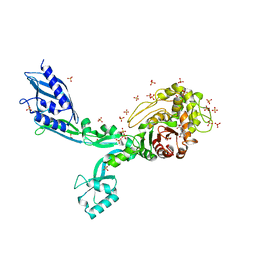 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S466 insertion variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3Z
 
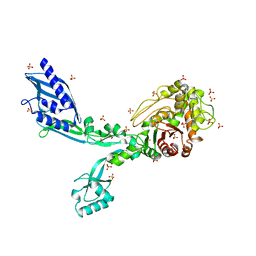 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) S422A variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8F3G
 
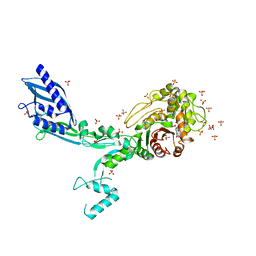 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M variant in the penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
2YGY
 
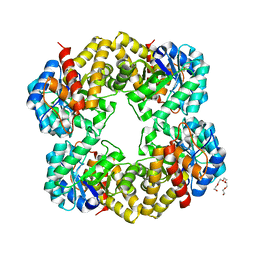 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form II | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL | | Authors: | Campeotto, I, Nelson, A, Berry, A, Phillips, S.E.V, Pearson, A.R. | | Deposit date: | 2011-04-23 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathological macromolecular crystallographic data affected by twinning, partial-disorder and exhibiting multiple lattices for testing of data processing and refinement tools.
Sci Rep, 8, 2018
|
|
7NDT
 
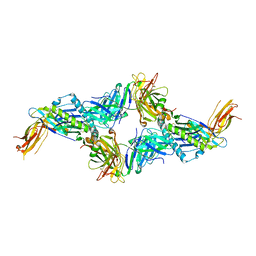 | | UL40:01 TCR in complex with HLA-E with a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7NDU
 
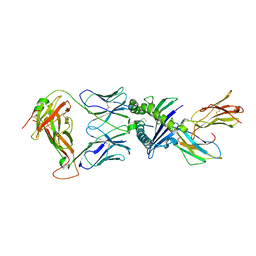 | | Gag:02 TCR in complex with HLA-E featuring a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, Gag6V(276-284 H4C), HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
8A5W
 
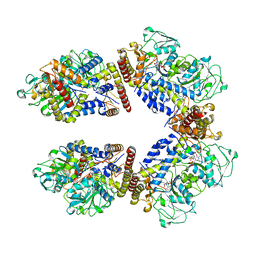 | | Crystal structure of the human phosphoserine aminotransferase (PSAT) in complex with O-phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, PHOSPHOSERINE, ... | | Authors: | Costanzi, E, Demitri, N, Ullah, R, Marchesan, F, Peracchi, A, Zangelmi, E, Storici, P, Campanini, B. | | Deposit date: | 2022-06-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | L-serine biosynthesis in the human central nervous system: Structure and function of phosphoserine aminotransferase.
Protein Sci., 32, 2023
|
|
7NDQ
 
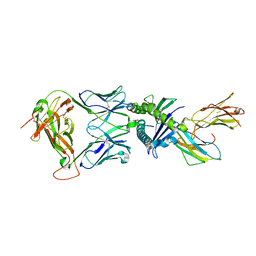 | | Gag:02 TCR in complex with HLA-E. | | Descriptor: | Beta-2-microglobulin, Gag6V, HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
2KK7
 
 | |
6BRW
 
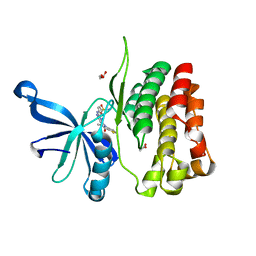 | | JAK2 JH2 in complex with XMU-MP-1 | | Descriptor: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, ACETATE ION, GLYCEROL, ... | | Authors: | Puleo, D.E, Schlessinger, J. | | Deposit date: | 2017-12-01 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | JAK2 JH2 Binders
To Be Published
|
|
6OZ6
 
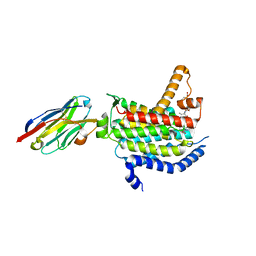 | | Crystal structure of MraY bound to 3'-hydroxymureidomycin A | | Descriptor: | (2~{S})-2-[[(2~{S})-1-[[(2~{S},3~{S})-3-[[(2~{S})-2-azanyl-3-(3-hydroxyphenyl)propanoyl]-methyl-amino]-1-[[(~{Z})-[(3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-ylidene]methyl]amino]-1-oxidanylidene-butan-2-yl]amino]-4-methylsulfanyl-1-oxidanylidene-butan-2-yl]carbamoylamino]-3-(3-hydroxyphenyl)propanoic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
8FE8
 
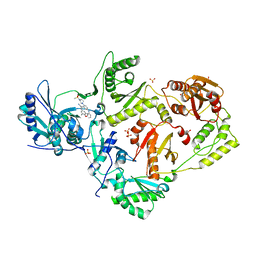 | | Crystal Structure of HIV-1 RT in Complex with the non-nucleoside inhibitor 18b1 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}pyrimidin-2-yl)amino]-2-[4-(methanesulfonyl)piperazin-1-yl]benzonitrile, Reverse transcriptase p51, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of diarylpyrimidine derivatives bearing piperazine sulfonyl as potent HIV-1 nonnucleoside reverse transcriptase inhibitors.
Commun Chem, 6, 2023
|
|
8IWH
 
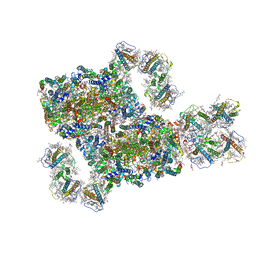 | | Structure and characteristics of a photosystem II supercomplex containing monomeric LHCX and dimeric FCPII antennae from the diatom Thalassiosira pseudonana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Feng, Y, Li, Z.H, Wang, W.D, Shen, J.R. | | Deposit date: | 2023-03-30 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Sci Adv, 9, 2023
|
|
6EWK
 
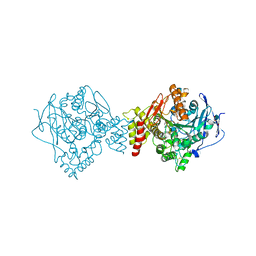 | | T. californica AChE in complex with a 3-hydroxy-2-pyridine aldoxime. | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-(5-morpholin-4-ylpentyl)pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | de la Mora, E, Weik, M, Braiki, A, Mougeot, R, Jean, L, Renard, P.I. | | Deposit date: | 2017-11-04 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Potent 3-Hydroxy-2-Pyridine Aldoxime Reactivators of Organophosphate-Inhibited Cholinesterases with Predicted Blood-Brain Barrier Penetration.
Chemistry, 24, 2018
|
|
8J5K
 
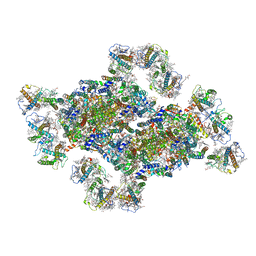 | | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2023-04-23 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
5UFM
 
 | | Crystal structure of Burkholderia thailandensis 1,6-didemethyltoxoflavin-N1-methyltransferase with bound 1,6-didemethyltoxoflavin and S-adenosylhomocysteine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Methyltransferase domain protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Fenwick, M.K, Ealick, S.E, Almabruk, K.H, Begley, T.P, Philmus, B. | | Deposit date: | 2017-01-05 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Biochemical Characterization and Structural Basis of Reactivity and Regioselectivity Differences between Burkholderia thailandensis and Burkholderia glumae 1,6-Didesmethyltoxoflavin N-Methyltransferase.
Biochemistry, 56, 2017
|
|
8J6N
 
 | | Crystal structure of Cystathionine gamma-lyase in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, Cystathionine gamma-lyase, GLYCEROL, ... | | Authors: | Hibi, R, Toma-Fukai, S, Shimizu, T, Hanaoka, K. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a cystathionine gamma-lyase (CSE) selective inhibitor targeting active-site pyridoxal 5'-phosphate (PLP) via Schiff base formation.
Sci Rep, 13, 2023
|
|
6MFP
 
 | | Crystal Structure of the RV305 C1-C2 specific ADCC potent antibody DH677.3 Fab in complex with HIV-1 clade A/E gp120 and M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Tolbert, W.D, Young, B, Pazgier, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Boosting with AIDSVAX B/E Enhances Env Constant Region 1 and 2 Antibody-Dependent Cellular Cytotoxicity Breadth and Potency.
J.Virol., 94, 2020
|
|
8B0N
 
 | | Cryo-EM structure of apolipoprotein N-acyltransferase Lnt from E. coli in complex with Lyso-PE | | Descriptor: | Apolipoprotein N-acyltransferase, [(2~{S})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-oxidanyl-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Degtjarik, O, Smithers, L, Boland, C, Caffrey, M, Shalev Benami, M. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
5E5I
 
 | | Structure of the ornithine aminotransferase from Toxoplasma gondii in complex with inactivator | | Descriptor: | 4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-4-enoic acid, 6-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]-4-oxidanylidene-hexanoic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the ornithine aminotransferase from Toxoplasma gondii in complex with inactivator.
To Be Published
|
|
5E3K
 
 | | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with (S)-4-amino-5-fluoropentanoic acid | | Descriptor: | 4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-4-enoic acid, Aminotransferase, CARBONATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-02 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with (S)-4-amino-5-fluoropentanoic acid
To Be Published
|
|
8RMY
 
 | |
