6GCI
 
 | | Structure of the bongkrekic acid-inhibited mitochondrial ADP/ATP carrier | | Descriptor: | Bongkrekic acid, CARDIOLIPIN, HEXAETHYLENE GLYCOL, ... | | Authors: | Ruprecht, J.J, King, M.S, Pardon, E, Aleksandrova, A.A, Zogg, T, Crichton, P.G, Steyaert, J, Kunji, E.R.S. | | Deposit date: | 2018-04-17 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier.
Cell, 176, 2019
|
|
8G61
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (AC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
5NQ4
 
 | | Cytotoxin-1 in DPC-micelle | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Volynsky, P.E, Pustovalova, Y.E, Konshina, A.G, Utkin, Y.N, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-13 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Impact of membrane partitioning on the spatial structure of an S-type cobra cytotoxin.
J. Biomol. Struct. Dyn., 36, 2018
|
|
5EEZ
 
 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 14.2 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6UBY
 
 | | Isolated cofilin bound to an actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-09-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7B3Y
 
 | | Structure of a nanoparticle for a COVID-19 vaccine candidate | | Descriptor: | Fibronectin binding protein,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Duyvesteyn, H.M.E, Stuart, D.I. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses.
Nat Commun, 12, 2021
|
|
7B6M
 
 | | Crystal structure of MurE from E.coli in complex with Z1198948504 | | Descriptor: | 1,2-ETHANEDIOL, N-[2-(2,5-Dioxopyrrolidin-1-yl)ethyl]-3-methylbenzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC), Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
8OH2
 
 | | Structure of the Tau-PAM4 Type 1 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
7B60
 
 | | Crystal structure of MurE from E.coli in complex with Z1269139261 | | Descriptor: | 4-[(furan-2-yl)methyl]-1lambda~6~,4-thiazinane-1,1-dione, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
8GI1
 
 | | Homo-octamer of PbuCsx28 protein | | Descriptor: | Accessory protein Csx28 | | Authors: | Park, J.U, Kellogg, E.H. | | Deposit date: | 2023-03-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Csx28 is a membrane pore that enhances CRISPR-Cas13b-dependent antiphage defense.
Science, 380, 2023
|
|
1H8G
 
 | | C-terminal domain of the major autolysin (C-LytA) from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, MAJOR AUTOLYSIN | | Authors: | Fernandez-Tornero, C, Garcia, E, Lopez, R, Gimenez-Gallego, G, Romero, A. | | Deposit date: | 2001-02-06 | | Release date: | 2002-01-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Novel Solenoid Fold in the Cell Wall Anchoring Domain of the Pneumococcal Virulence Factor Lyta
Nat.Struct.Biol., 8, 2001
|
|
7TBU
 
 | | Crystal structure of the 5-enolpyruvate-shikimate-3-phosphate synthase (EPSPS) domain of Aro1 from Candida albicans in complex with shikimate-3-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-enolpyruvylshikimate-3-phosphate synthase, SHIKIMATE-3-PHOSPHATE | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
7TBV
 
 | | Crystal structure of the shikimate kinase + 3-dehydroquinate dehydratase + 3-dehydroshikimate dehydrogenase domains of Aro1 from Candida albicans | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
7TQL
 
 | | CryoEM structure of the human 40S small ribosomal subunit in complex with translation initiation factors eIF1A and eIF5B. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Lapointe, C.P, Grosely, R, Sokabe, M, Alvarado, C, Wang, J, Montabana, E, Villa, N, Shin, B, Dever, T, Fraser, C, Fernandez, I.S, Puglisi, J.D. | | Deposit date: | 2022-01-26 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | eIF5B and eIF1A reorient initiator tRNA to allow ribosomal subunit joining.
Nature, 607, 2022
|
|
5NJH
 
 | | Triazolopyrimidines stabilize microtubules by binding to the vinca inhibitor site of tubulin | | Descriptor: | 5-chloranyl-7-[(1~{R},5~{S})-3-methoxy-8-azabicyclo[3.2.1]octan-8-yl]-6-[2,4,6-tris(fluoranyl)phenyl]-[1,2,4]triazolo[1,5-a]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sharma, A, Calvo, G.S, Prota, A.E, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2017-03-28 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Triazolopyrimidines Are Microtubule-Stabilizing Agents that Bind the Vinca Inhibitor Site of Tubulin.
Cell Chem Biol, 24, 2017
|
|
7AHL
 
 | | ALPHA-HEMOLYSIN FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | ALPHA-HEMOLYSIN | | Authors: | Song, L, Hobaugh, M, Shustak, C, Cheley, S, Bayley, H, Gouaux, J.E. | | Deposit date: | 1996-12-02 | | Release date: | 1998-01-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore.
Science, 274, 1996
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
6BTL
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor RD117 1-[2-(Piperazin-1-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole | | Descriptor: | 1-[2-(piperazin-1-yl)ethyl]-5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bryson, S, De Gasparo, R, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2017-12-06 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Biological Evaluation and X-ray Co-crystal Structures of Cyclohexylpyrrolidine Ligands for Trypanothione Reductase, an Enzyme from the Redox Metabolism of Trypanosoma.
ChemMedChem, 13, 2018
|
|
5EEV
 
 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 3.88 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6TGG
 
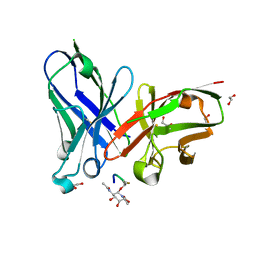 | | scFv-1SM3 in complex with glycopeptide containing an sp2-imino sugar | | Descriptor: | 1,2-ETHANEDIOL, Mucin-1, ScFv_SM3, ... | | Authors: | Bermejo, I.A, Navo, C.D, Castro-Lopez, J, Samchez-Fernandez, E.M, Guerreiro, A, Avenoza, A, Busto, J.H, Garcia-Martin, F, Nishimura, S.I, Garcia-Fernandez, J.M, Ortiz-Mellet, C, Bernardes, G.J.L, Hurtado-Guerrero, R, Peregrina, J.M, Corzana, F. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, conformational analysis and in vivo assays of an anti-cancer vaccine that features an unnatural antigen based on an sp 2 -iminosugar fragment.
Chem Sci, 11, 2020
|
|
7B6N
 
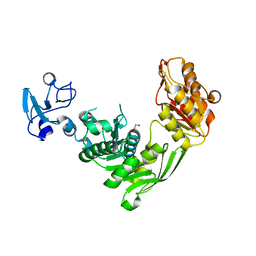 | | Crystal structure of MurE from E.coli in complex with Z757284952 | | Descriptor: | 1,2-ETHANEDIOL, N-(2-(2,4-dioxothiazolidin-3-yl)ethyl)-3-methylbenzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
5D2R
 
 | |
6UIT
 
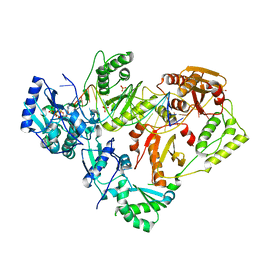 | | HIV-1 wild-type reverse transcriptase-DNA complex with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA primer, DNA template, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2019-10-01 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.80607414 Å) | | Cite: | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
5EF0
 
 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 16.7 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1NUJ
 
 | | THE LEADZYME STRUCTURE BOUND TO MG(H20)6(II) AT 1.8 A RESOLUTION | | Descriptor: | 5'-R(*CP*GP*GP*AP*CP*CP*GP*AP*GP*CP*CP*AP*G)-3', 5'-R(*GP*CP*UP*GP*GP*GP*AP*GP*UP*CP*C)-3', MAGNESIUM ION | | Authors: | Wedekind, J.E, Mckay, D.B. | | Deposit date: | 2003-01-31 | | Release date: | 2003-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the leadzyme at 1.8 A resolution: metal ion binding and the implications
for catalytic mechanism and allo site ion regulation.
BIOCHEMISTRY, 42, 2003
|
|
