5LZT
 
 | | Structure of the mammalian ribosomal termination complex with eRF1 and eRF3. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZD
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the GTPase activated state (GA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
7D1Z
 
 | | Cryo-EM structure of SET8-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
8IEJ
 
 | | RNF20-RNF40/hRad6A-Ub/nucleosome complex | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Ai, H, Deng, Z, Sun, M, Du, Y, Pan, M, Liu, L. | | Deposit date: | 2023-02-15 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.
Mol.Cell, 83, 2023
|
|
5JKW
 
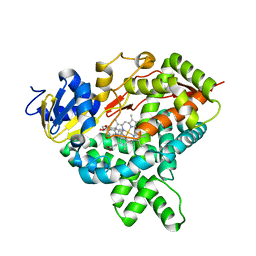 | |
8IEG
 
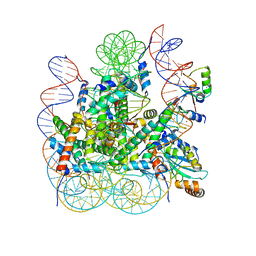 | | Bre1(mRBD-RING)/Rad6-Ub/nucleosome complex | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase BRE1, Histone H2A type 1-B/E, ... | | Authors: | Ai, H, Deng, Z, Pan, M, Liu, L. | | Deposit date: | 2023-02-15 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.
Mol.Cell, 83, 2023
|
|
5J9T
 
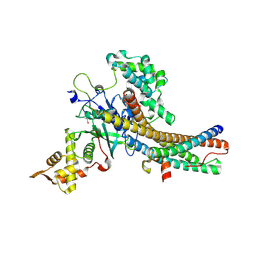 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
7D0J
 
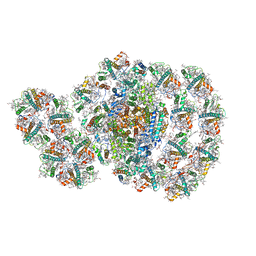 | | Photosystem I-LHCI-LHCII of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wang, W.D, Shen, L.L, Huang, Z.H, Han, G.Y, Zhang, X, Shen, J.R. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure of photosystem I-LHCI-LHCII from the green alga Chlamydomonas reinhardtii in State 2.
Nat Commun, 12, 2021
|
|
8JDJ
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Asp(Q34) and mRNA(GAU) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDK
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Asp(ManQ34) and mRNA(GAU) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
5SZ9
 
 | | Structure-based design of a new series of N-piperidin-3-ylpyrimidine-5-carboxamides as renin inhibitors | | Descriptor: | (azepan-1-yl)(2-{[(furan-2-yl)methyl]amino}-6-methylpyridin-3-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.C, Lane, W. | | Deposit date: | 2016-08-12 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based design of a new series of N-(piperidin-3-yl)pyrimidine-5-carboxamides as renin inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5FLM
 
 | | Structure of transcribing mammalian RNA polymerase II | | Descriptor: | DNA, DNA-RNA ELONGATION SCAFFOLD, DNA-DIRECTED RNA POLYMERASE, ... | | Authors: | Bernecky, C, Herzog, F, Baumeister, W, Plitzko, J.M, Cramer, P. | | Deposit date: | 2015-10-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of Transcribing Mammalian RNA Polymerase II
Nature, 529, 2016
|
|
5SXN
 
 | | Structure-based design of a new series of N-piperidin-3-ylpyrimidine-5-carboxamides as renin inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.-C, Lane, W. | | Deposit date: | 2016-08-09 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of a new series of N-(piperidin-3-yl)pyrimidine-5-carboxamides as renin inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5T2C
 
 | | CryoEM structure of the human ribosome at 3.6 Angstrom resolution | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Zhang, X, Lai, M, Zhou, Z.H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-01-25 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and stabilization of kinetoplastid-specific split rRNAs revealed by comparing leishmanial and human ribosomes.
Nat Commun, 7, 2016
|
|
5LZE
 
 | | Structure of the 70S ribosome with Sec-tRNASec in the classical pre-translocation state (C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5LZC
 
 | | Structure of SelB-Sec-tRNASec bound to the 70S ribosome in the codon reading state (CR) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
5DF5
 
 | | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution. | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | The structure of oxidized rat cytochrome c (T28E) at 1.30 angstroms resolution.
To Be Published
|
|
8K2C
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
7K1K
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex IV) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7JRP
 
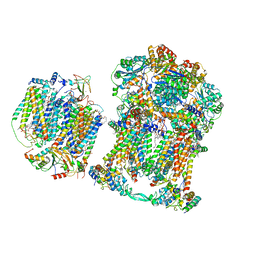 | | Plant Mitochondrial complex SC III2+IV from Vigna radiata | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Alpha-MPP, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of respiratory complex III 2 , complex IV, and supercomplex III 2 -IV from vascular plants.
Elife, 10, 2021
|
|
5LZX
 
 | | Structure of the mammalian rescue complex with Pelota and Hbs1l assembled on a UGA stop codon. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
5LZV
 
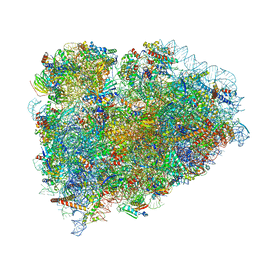 | | Structure of the mammalian ribosomal termination complex with accommodated eRF1(AAQ) and ABCE1. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
7K48
 
 | | Structure of NavAb/Nav1.7-VS2A chimera trapped in the resting state by tarantula toxin m3-Huwentoxin-IV | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Ion transport protein,Sodium channel protein type 9 subunit alpha chimera, Mu-theraphotoxin-Hs2a | | Authors: | Wisedchaisri, G, Tonggu, L, Gamal El-Din, T.M, McCord, E, Zheng, N, Catterall, W.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for High-Affinity Trapping of the Na V 1.7 Channel in Its Resting State by Tarantula Toxin.
Mol.Cell, 81, 2021
|
|
5IFL
 
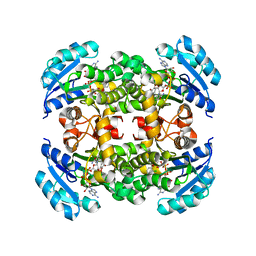 | | Crystal structure of B. pseudomallei FabI in complex with NAD and triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
8P25
 
 | | Solution structure of a chimeric U2AF2 RRM2 / FUBP1 N-Box | | Descriptor: | Splicing factor U2AF 65 kDa subunit,Far upstream element-binding protein 1 | | Authors: | Hipp, C, Sattler, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | FUBP1 is a general splicing factor facilitating 3' splice site recognition and splicing of long introns.
Mol.Cell, 83, 2023
|
|
