5I4H
 
 | |
2GU1
 
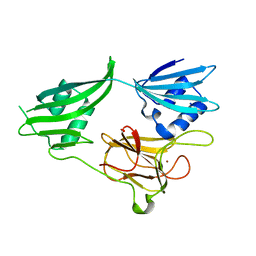 | | Crystal structure of a zinc containing peptidase from vibrio cholerae | | Descriptor: | SODIUM ION, ZINC ION, Zinc peptidase | | Authors: | Sugadev, R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-28 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative lysostaphin peptidase from Vibrio cholerae.
Proteins, 72, 2008
|
|
2GU9
 
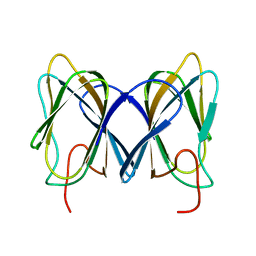 | |
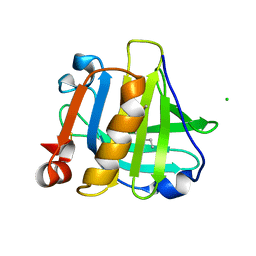
5IO7
 
 | |
7S25
 
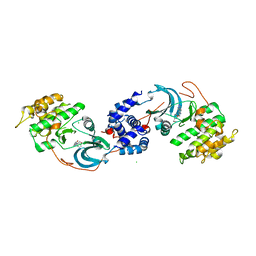 | | ROCK1 IN COMPLEX WITH LIGAND G4998 | | Descriptor: | 2-[3-(methoxymethyl)phenyl]-N-[4-(1H-pyrazol-4-yl)phenyl]acetamide, CHLORIDE ION, Rho-associated protein kinase 1 | | Authors: | Ganichkin, O, Harris, S.F, Steinbacher, S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.337 Å) | | Cite: | Chemical space docking enables large-scale structure-based virtual screening to discover ROCK1 kinase inhibitors.
Nat Commun, 13, 2022
|
|
2RLP
 
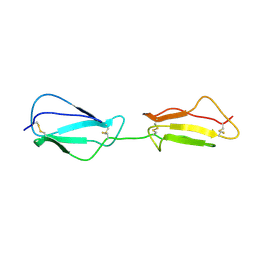 | | NMR structure of CCP modules 1-2 of complement factor H | | Descriptor: | Complement factor H | | Authors: | Hocking, H.G, Herbert, A.P, Pangburn, M.K, Kavanagh, D, Barlow, P.N, Uhrin, D. | | Deposit date: | 2007-07-28 | | Release date: | 2008-02-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations.
J.Biol.Chem., 283, 2008
|
|
2RMW
 
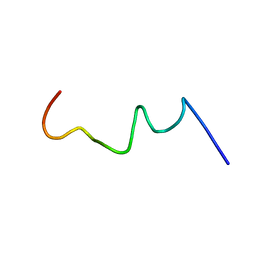 | |
2GR2
 
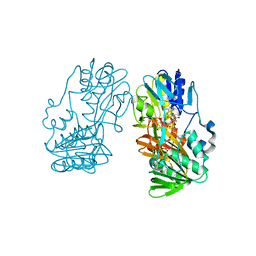 | |
5IOM
 
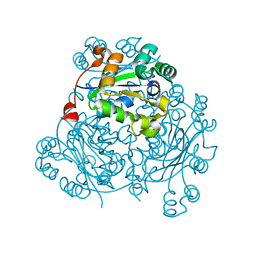 | | Crystal Structure of Nucleoside Diphosphate Kinase from Schistosoma mansoni is space group P6322 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Torini, J.R.S, Romanello, L, Bird, L.E, Nettleship, J.E, Owens, R.J, Aller, P, DeMarco, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-03-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a Schistosoma mansoni NDPK expressed in sexual and digestive organs.
Mol.Biochem.Parasitol., 2019
|
|
7D3C
 
 | |
2GRG
 
 | | Solution NMR Structure of Protein YNR034W-A from Saccharomyces cerevisiae. Northeast Structural Genomics Consortium Target YT727; Ontario Center for Structural Proteomics Target yst6499. | | Descriptor: | hypothetical protein; Ynr034w-ap | | Authors: | Wu, B, Yee, A, Ramelot, T, Lemak, A, Semesi, A, Kennedy, M, Edward, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-24 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein yst6499 from Saccharomyces cerevisiae/ Northeast Structural Genomics Consortium Target YT727/ Ontario Center for Structural Proteomics Target yst6499
To be Published
|
|
2GW3
 
 | | Crystal structure of stony coral fluorescent protein Kaede, green form | | Descriptor: | Kaede, NICKEL (II) ION | | Authors: | Hayashi, I, Mizuno, H, Miyawaki, A, Ikura, M. | | Deposit date: | 2006-05-03 | | Release date: | 2007-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic evidence for water-assisted photo-induced peptide cleavage in the stony coral fluorescent protein Kaede.
J.Mol.Biol., 372, 2007
|
|
7S4T
 
 | | Crystal structure of CDK2 liganded with compound EF2252 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(6-chloro-1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
2RNF
 
 | | X-RAY CRYSTAL STRUCTURE OF HUMAN RIBONUCLEASE 4 IN COMPLEX WITH D(UP) | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, RIBONUCLEASE 4 | | Authors: | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 1998-11-03 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
2GWF
 
 | | Structure of a USP8-NRDP1 complex | | Descriptor: | RING finger protein 41, Ubiquitin carboxyl-terminal hydrolase 8 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amino-terminal Dimerization, NRDP1-Rhodanese Interaction, and Inhibited Catalytic Domain Conformation of the Ubiquitin-specific Protease 8 (USP8).
J.Biol.Chem., 281, 2006
|
|
2RNL
 
 | | Solution structure of the EGF-like domain from human Amphiregulin | | Descriptor: | Amphiregulin | | Authors: | Qin, X, Hayashi, F, Terada, T, Shirouzu, M, Watanabe, S, Kigawa, T, Yabuta, N, Nojima, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the EGF-like domain from human Amphiregulin
To be Published
|
|
2GSF
 
 | | The Human Epha3 Receptor Tyrosine Kinase and Juxtamembrane Region | | Descriptor: | Ephrin type-A receptor 3 | | Authors: | Walker, J.R, Davis, T, Dong, A, Newman, E.M, MacKenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-26 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure Of The Human Epha3 Receptor Tyrosine Kinase and Juxtamembrane Region
To be Published
|
|
7SA2
 
 | |
2GX1
 
 | | Solution structure and alanine scan of a spider toxin that affects the activation of mammalian sodium channels | | Descriptor: | Neurotoxin magi-5 | | Authors: | Sabo, J.K, Corzo, G, Bosmans, F, Billen, B, Villegas, E, Tytgat, J, Norton, R.S. | | Deposit date: | 2006-05-08 | | Release date: | 2006-12-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Alanine Scan of a Spider Toxin That Affects the Activation of Mammalian Voltage-gated Sodium Channels
J.Biol.Chem., 282, 2007
|
|
2GXS
 
 | |
2RNX
 
 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the HUman Transcriptional Co-Activators PCAF and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
7S26
 
 | | ROCK1 IN COMPLEX WITH LIGAND G5018 | | Descriptor: | 2-[methyl(phenyl)amino]-1-[4-(1H-pyrrolo[2,3-b]pyridin-3-yl)-3,6-dihydropyridin-1(2H)-yl]ethan-1-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Rho-associated protein kinase 1 | | Authors: | Ganichkin, O, Harris, S.F, Steinbacher, S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Chemical space docking enables large-scale structure-based virtual screening to discover ROCK1 kinase inhibitors.
Nat Commun, 13, 2022
|
|
2RON
 
 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
7S4E
 
 | |
2RPQ
 
 | | Solution Structure of a SUMO-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3 | | Descriptor: | Activating transcription factor 7-interacting protein 1, Small ubiquitin-related modifier 2 | | Authors: | Sekiyama, N, Ikegami, T, Yamane, T, Ikeguchi, M, Uchimura, Y, Baba, D, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2008-07-07 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3
J.Biol.Chem., 283, 2008
|
|
