3ZWZ
 
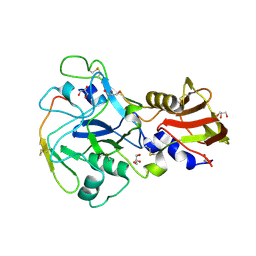 | | Crystal structure of Plasmodium falciparum AMA1 in complex with a 39aa PfRON2 peptide | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, AMA1, GLYCEROL, ... | | Authors: | Vulliez-Le Normand, B, Tonkin, M.L, Lamarque, M.H, Langer, S, Hoos, S, Roques, M, Saul, F.A, Faber, B.W, Bentley, G.A, Boulanger, M.J, Lebrun, M. | | Deposit date: | 2011-08-03 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Insight Into the Malaria Parasite Moving Junction Complex
Plos Pathog., 8, 2012
|
|
8G2Z
 
 | | 48-nm doublet microtubule from Tetrahymena thermophila strain CU428 | | Descriptor: | B2B3_fMIP, B5B6_fMIP, CFAM166A, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins.
Nat Commun, 14, 2023
|
|
8G3D
 
 | | 48-nm doublet microtubule from Tetrahymena thermophila strain K40R | | Descriptor: | B2B3_fMIP, B5B6_fMIP, CFAM166A, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins.
Nat Commun, 14, 2023
|
|
2IXY
 
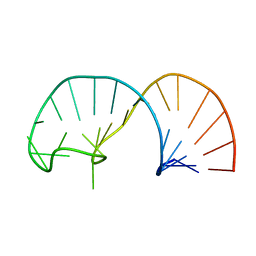 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | Descriptor: | 5'-R(*GP*GP*CP*CP*UP*CP*CP*AP*AP*GP *CP*UP*GP*UP*GP*CP*CP*UP*UP*GP*GP*GP*UP*GP*GP*CP*C)-3' | | Authors: | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
8UVK
 
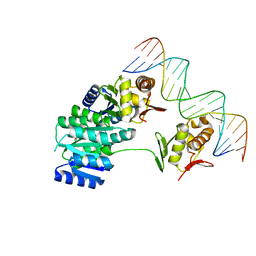 | | CosR DNA bound form II | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*T)-3'), ... | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UVX
 
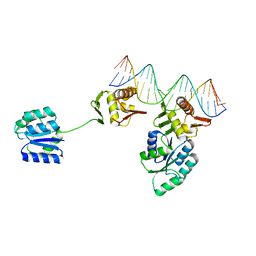 | | CosR DNA bound form I | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
2JMK
 
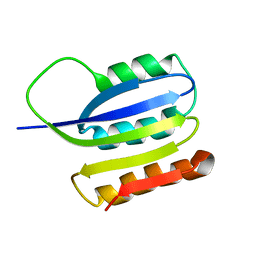 | | Solution structure of ta0956 | | Descriptor: | Hypothetical protein Ta0956 | | Authors: | Koo, B, Jung, J, Jung, H, Nam, H, Kim, Y, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2006-11-20 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical novel-fold protein TA0956 from Thermoplasma acidophilum
Proteins, 69, 2007
|
|
6F3C
 
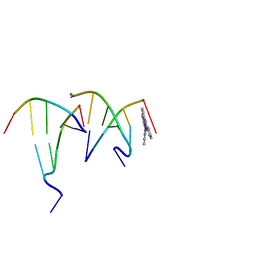 | | The cytotoxic [Pt(H2bapbpy)] platinum complex interacting with the CGTACG hexamer | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*G)-3'), MAGNESIUM ION, [Pt(H2bapbpy)] platinum | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Papi, F. | | Deposit date: | 2017-11-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Induction of a Four-Way Junction Structure in the DNA Palindromic Hexanucleotide 5'-d(CGTACG)-3' by a Mononuclear Platinum Complex.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6PPZ
 
 | | Crystal structure of NeuB, an N-acetylneuraminate synthase from Neisseria meningitidis, in complex with manganese, inorganic phosphate, and N-acetylmannosamine (NeuB.Mn2+.Pi.ManNAc) | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, MANGANESE (II) ION, N-acetylneuraminate synthase, ... | | Authors: | Berti, P.J, Junop, M.S. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | NeuNAc Oxime: A Slow-Binding and Effectively Irreversible Inhibitor of the Sialic Acid Synthase NeuB.
Biochemistry, 58, 2019
|
|
5H1N
 
 | |
5H2D
 
 | | Crystal structure of Osh1 ORD domain in complex with ergosterol | | Descriptor: | ERGOSTEROL, KLLA0C04147p, SULFATE ION | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
4Z90
 
 | | ELIC bound with the anesthetic isoflurane in the resting state | | Descriptor: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Chen, Q, Kinde, M, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
5H28
 
 | |
5GYY
 
 | | Plant receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S-locus protein 11, S-receptor kinase SRK9 | | Authors: | Ma, R, Han, Z, Hu, Z, Lin, G, Gong, X, Zhang, H, June, N, Chai, J. | | Deposit date: | 2016-09-24 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Plant receptor complex at 2.35 Angstroms resolution
To Be Published
|
|
5H2A
 
 | |
8D3Q
 
 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/NoPAM prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
8D3M
 
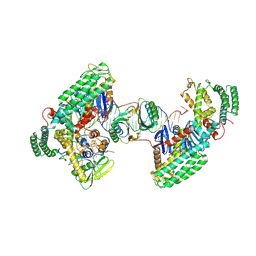 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/Processed prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
8D3L
 
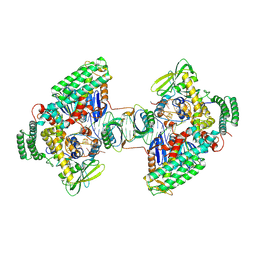 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/PAM prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
8D3P
 
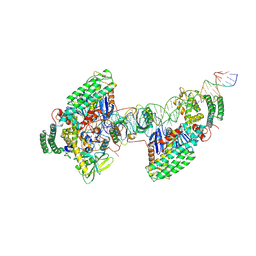 | | Type I-C Cas4-Cas1-Cas2 complex bound to half-site integration intermediate (HSI) | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
4Z91
 
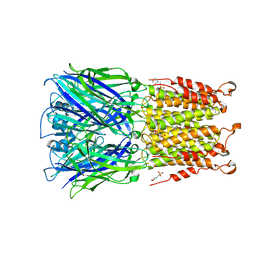 | | ELIC cocrystallized with isofluorane in a desensitized state | | Descriptor: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 1.7.6 3-bromanylpropan-1-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Chen, Q, Kinde, M.N, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3915 Å) | | Cite: | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
4ZTT
 
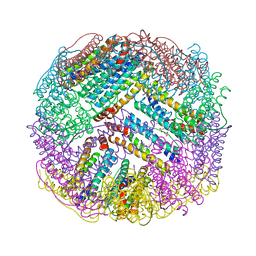 | | Crystal structures of ferritin mutants reveal diferric-peroxo intermediates | | Descriptor: | Bacterial non-heme ferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Kim, S, Park, Y.H, Jung, S.W, Seok, J.H, Chung, Y.B, Lee, D.B, Gowda, G, Lee, J.H, Han, H.R, Cho, A.E, Lee, C, Chung, M.S, Kim, K.H. | | Deposit date: | 2015-05-15 | | Release date: | 2016-06-15 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
4K1O
 
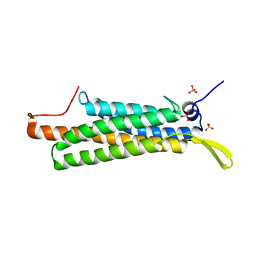 | |
5A3K
 
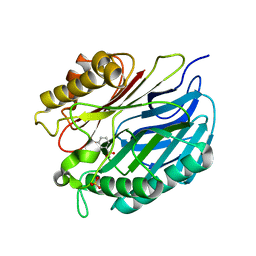 | | Chorismatase mechanisms reveal fundamentally different types of reaction in a single conserved protein fold | | Descriptor: | 3-HYDROXYBENZOIC ACID, PUTATIVE PTERIDINE-DEPENDENT DIOXYGENASE, SULFATE ION | | Authors: | Hubrich, F, Juneja, P, Mueller, M, Diederichs, K, Welte, W, Andexer, J.N. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | Chorismatase Mechanisms Reveal Fundamentally Different Types of Reaction in a Single Conserved Protein Fold.
J.Am.Chem.Soc., 137, 2015
|
|
8E0Z
 
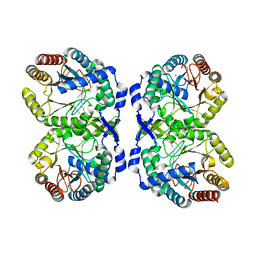 | |
8E0Y
 
 | | DAHP (3-deoxy-D-arabinoheptulosonate-7-phosphate) Synthase complexed with DAHP oxime, Pr(III), and Pi in unbound:(bound)2:other Conformations | | Descriptor: | ACETATE ION, CITRATE ANION, DAHP Oxime, ... | | Authors: | Berti, P.J, Junop, M.S, Grainger, R. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Role of Half-of-Sites Reactivity and Inter-Subunit Communications in DAHP Synthase Catalysis and Regulation.
Biochemistry, 61, 2022
|
|
