3A02
 
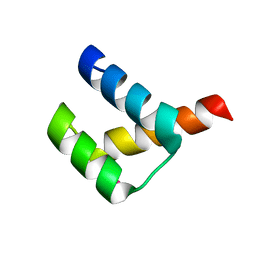 | | Crystal structure of Aristaless homeodomain | | Descriptor: | CADMIUM ION, CHLORIDE ION, Homeobox protein aristaless | | Authors: | Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
3A0V
 
 | | PAS domain of histidine kinase ThkA (TM1359) (SeMet, F486M/F489M) | | Descriptor: | ETHANOL, Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3BFU
 
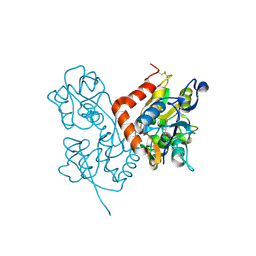 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | Descriptor: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
3A18
 
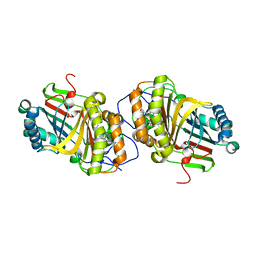 | | Crystal Structure of Aldoxime Dehydratase (OxdRE) in Complex with Butyraldoxime (soaked crystal) | | Descriptor: | (1Z)-butanal oxime, Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3N5U
 
 | | Crystal structure of an Rb C-terminal peptide bound to the catalytic subunit of PP1 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Retinoblastoma-associated protein, ... | | Authors: | Hirschi, A.M, Cecchini, M, Steinhardt, R.C, Dick, F.A, Rubin, S.M. | | Deposit date: | 2010-05-25 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7WBI
 
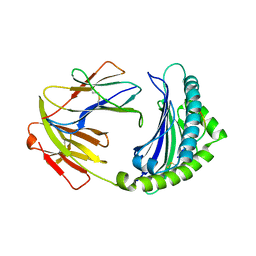 | | BF2*1901-FLU | | Descriptor: | Beta-2-microglobulin, ILE-ARG-HIS-GLU-ASN-ARG-MET-VAL-LEU, MHC class I alpha chain 2 | | Authors: | Liu, W.J. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Wider and Deeper Peptide-Binding Groove for the Class I Molecules from B15 Compared with B19 Chickens Correlates with Relative Resistance to Marek's Disease.
J Immunol., 210, 2023
|
|
3A4U
 
 | | Crystal structure of MCFD2 in complex with carbohydrate recognition domain of ERGIC-53 | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Nishio, M, Kamiya, Y, Mizushima, T, Wakatsuki, S, Sasakawa, H, Yamamoto, K, Uchiyama, S, Noda, M, McKay, A.R, Fukui, K, Hauri, H.P, Kato, K. | | Deposit date: | 2009-07-17 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3A8W
 
 | | Crystal Structure of PKCiota kinase domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein kinase C iota type, SULFATE ION | | Authors: | Takimura, T, Kamata, K. | | Deposit date: | 2009-10-11 | | Release date: | 2010-05-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the PKC-iota kinase domain in its ATP-bound and apo forms reveal defined structures of residues 533-551 in the C-terminal tail and their roles in ATP binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7WIK
 
 | |
3BOK
 
 | |
3A07
 
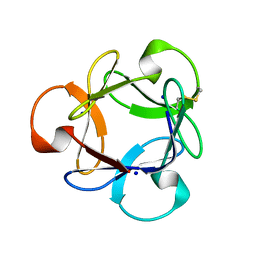 | | Crystal Structure of Actinohivin; Potent anti-HIV Protein | | Descriptor: | Actinohivin, SODIUM ION | | Authors: | Tsunoda, M, Suzuki, K, Sagara, T, Takenaka, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-08-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Mechanism by which the lectin actinohivin blocks HIV infection of target cells
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3NF3
 
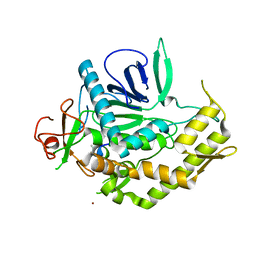 | | Crystal structure of BoNT/A LC with JTH-NB-7239 peptide | | Descriptor: | BoNT/A, JTH-NB72-39 inhibitor, NICKEL (II) ION, ... | | Authors: | Zuniga, J.E. | | Deposit date: | 2010-06-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Iterative structure-based peptide-like inhibitor design against the botulinum neurotoxin serotype A.
Plos One, 5, 2010
|
|
3AED
 
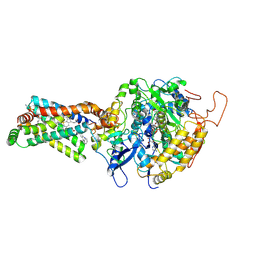 | | Crystal structure of porcine heart mitochondrial complex II bound with 2-Iodo-N-phenyl-benzamide | | Descriptor: | 2-iodo-N-phenylbenzamide, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II bound with 2-Iodo-N-phenyl-benzamide
To be Published
|
|
7WCA
 
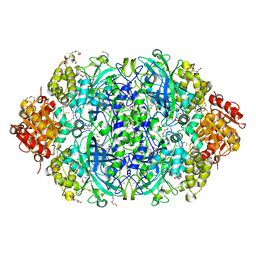 | | CATPO mutant - E484A | | Descriptor: | CALCIUM ION, Catalase, PENTAETHYLENE GLYCOL, ... | | Authors: | Yuzugullu Karakus, Y, Balci Unver, S, Zengin Karatas, M, Goc, G, Pearson, A.R, Yorke, B. | | Deposit date: | 2021-12-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Investigation of how gate residues in the main channel affect the catalytic activity of Scytalidium thermophilum catalase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
3BXM
 
 | | Structure of an inactive mutant of human glutamate carboxypeptidase II [GCPII(E424A)] in complex with N-acetyl-Asp-Glu (NAAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lubkowski, J, Barinka, C. | | Deposit date: | 2008-01-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Reaction mechanism of glutamate carboxypeptidase II revealed by mutagenesis, X-ray crystallography, and computational methods.
Biochemistry, 48, 2009
|
|
3C45
 
 | | Human dipeptidyl peptidase IV/CD26 in complex with a fluoroolefin inhibitor | | Descriptor: | (2S,3S)-3-{3-[2-chloro-4-(methylsulfonyl)phenyl]-1,2,4-oxadiazol-5-yl}-1-cyclopentylidene-4-cyclopropyl-1-fluorobutan-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Edmondson, S.D, Weber, A.E. | | Deposit date: | 2008-01-29 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Fluoroolefins as amide bond mimics in dipeptidyl peptidase IV inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3M37
 
 | |
3BYA
 
 | |
3CCC
 
 | | Crystal Structure of Human DPP4 in complex with a benzimidazole derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(aminomethyl)-6-(2-chlorophenyl)-1-methyl-1H-benzimidazole-5-carbonitrile, ... | | Authors: | Wallace, M.B, Skene, R.J. | | Deposit date: | 2008-02-25 | | Release date: | 2008-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure-based design and synthesis of benzimidazole derivatives as dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3N2G
 
 | | TUBULIN-NSC 613863: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Barbier, P, Dorleans, A, Devred, F, Sanz, L, Allegro, D, Alfonso, C, Knossow, M, Peyrot, V, Andreu, J.M. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Stathmin and interfacial microtubule inhibitors recognize a naturally curved conformation of tubulin dimers.
J.Biol.Chem., 285, 2010
|
|
3NBI
 
 | | Crystal structure of human RMI1 N-terminus | | Descriptor: | RecQ-mediated genome instability protein 1 | | Authors: | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
3A0K
 
 | | Crystal structure of an antiflamatory legume lectin from Cymbosema roseum seeds | | Descriptor: | ALPHA-AMINOBUTYRIC ACID, CALCIUM ION, Cymbosema roseum mannose-specific lectin, ... | | Authors: | Rocha, B.A.M, Delatorre, P, Marinho, E.S, Benevides, R.G, Moura, T.R, Souza, L.A.G, Nascimento, K.S, Sampaio, A.H, Cavada, B.S. | | Deposit date: | 2009-03-20 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for both pro- and anti-inflammatory response induced by mannose-specific legume lectin from Cymbosema roseum
Biochimie, 2011
|
|
3NKE
 
 | | High resolution structure of the C-terminal domain CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, SULFITE ION, ... | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
3A46
 
 | | Crystal structure of MvNei1/THF complex | | Descriptor: | DNA, Formamidopyrimidine-DNA glycosylase, GLYCEROL | | Authors: | Imamura, K, Wallace, S, Doublie, S. | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of a Viral NEIL1 Ortholog Unliganded and Bound to Abasic Site-containing DNA
J.Biol.Chem., 284, 2009
|
|
3A45
 
 | | Crystal structure of MvNei1_2 | | Descriptor: | Formamidopyrimidine-DNA glycosylase, POTASSIUM ION | | Authors: | Imamura, K, Wallace, S, Doublie, S. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of a Viral NEIL1 Ortholog Unliganded and Bound to Abasic Site-containing DNA
J.Biol.Chem., 284, 2009
|
|
