3CZA
 
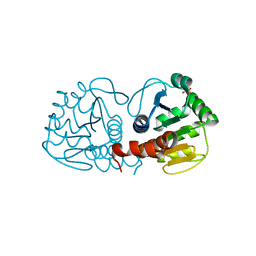 | | Crystal Structure of E18D DJ-1 | | Descriptor: | MALONIC ACID, Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hashim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
3D2Q
 
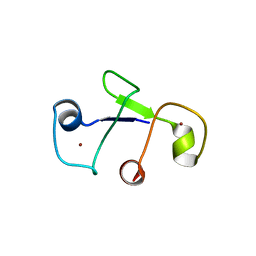 | |
3LK4
 
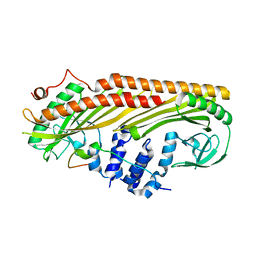 | | Crystal structure of CapZ bound to the uncapping motif from CD2AP | | Descriptor: | CD2-associated protein, F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2 | | Authors: | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Cooper, J.A, Robinson, R.C. | | Deposit date: | 2010-01-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L6N
 
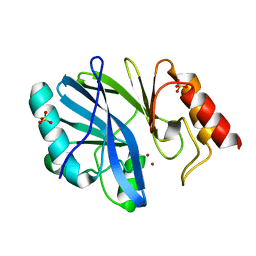 | |
3D0R
 
 | | Crystal structure of calG3 from Micromonospora echinospora determined in space group P2(1) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Protein CalG3 | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N. | | Deposit date: | 2008-05-02 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis.
Chem.Biol., 15, 2008
|
|
3D4L
 
 | | Human dipeptidyl peptidase IV/CD26 in complex with a novel inhibitor | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2008-05-14 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of new binding elements in DPP-4 inhibition and their applications in novel DPP-4 inhibitor design.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3D7H
 
 | |
3LK2
 
 | | Crystal structure of CapZ bound to the uncapping motif from CARMIL | | Descriptor: | F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2, Leucine-rich repeat-containing protein 16A | | Authors: | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Aguda, A.H, Larsson, M, Cooper, J.A, Robinson, R.C. | | Deposit date: | 2010-01-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L9P
 
 | |
7X45
 
 | | Grass carp interferon gamma related | | Descriptor: | Interferon gamma | | Authors: | Wang, J, Zou, J, Zhu, X. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-14 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Novel Dimeric Architecture of an IFN-gamma-Related Cytokine Provides Insights into Subfunctionalization of Type II IFNs in Teleost Fish.
J Immunol., 209, 2022
|
|
7XCE
 
 | | Crystal structure of Ankyrin G in complex with neurofascin | | Descriptor: | GLYCEROL, Neurofascin,Ankyrin-3, SULFATE ION | | Authors: | He, L, Li, J, Wang, C. | | Deposit date: | 2022-03-24 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Ankyrin-G in complex with a fragment of Neurofascin reveals binding mechanisms required for integrity of the axon initial segment.
J.Biol.Chem., 298, 2022
|
|
3LK3
 
 | | Crystal structure of CapZ bound to the CPI and CSI uncapping motifs from CARMIL | | Descriptor: | F-actin-capping protein subunit alpha-1, F-actin-capping protein subunit beta isoforms 1 and 2, Leucine-rich repeat-containing protein 16A | | Authors: | Hernandez-Valladares, M, Kim, T, Kannan, B, Tung, A, Cooper, J.A, Robinson, R.C. | | Deposit date: | 2010-01-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural characterization of a capping protein interaction motif defines a family of actin filament regulators.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3BFT
 
 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (S)-TDPA at 2.25 A resolution | | Descriptor: | (2S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
7WW4
 
 | |
7WWC
 
 | | Crystal structure of beta-1,3(4)-glucanase with Laminaritriose | | Descriptor: | beta-1,3(4)-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Jiang, Z.Q, Ma, J.W. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2021625 Å) | | Cite: | Crystal structure of beta-1,3(4)-glucanase with Laminaritriose
To Be Published
|
|
3L9Q
 
 | |
3BI0
 
 | |
3LCS
 
 | |
3BK9
 
 | | H55A mutant of tryptophan 2,3-dioxygenase from Xanthomonas campestris | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, Tryptophan 2,3-dioxygenase | | Authors: | Bruckmann, C, Mowat, C.G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling substrate binding
Biochemistry, 47, 2008
|
|
7UTT
 
 | |
7UUX
 
 | | ATP binds to Cyclic GMP AMP synthase (cGAS) through Mg coordination | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, MAGNESIUM ION, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7V0W
 
 | | Structure of Ternary Complex of cGAS with dsDNA and Bound 5 -pppG(2,5 )pA | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cyclic GMP-AMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-11 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
3LSF
 
 | | Piracetam bound to the ligand binding domain of GluA2 | | Descriptor: | 2-(2-oxopyrrolidin-1-yl)acetamide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Piracetam Defines a New Binding Site for Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazole-propionic Acid (AMPA) Receptors.
J.Med.Chem., 53, 2010
|
|
7UXW
 
 | | Structure of ATP and GTP bind to Cyclic GMP AMP synthase (cGAS) through Mg coordination | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
7UYQ
 
 | | Structure of GTP binds to Cyclic GMP AMP synthase (cGAS) through Mg coordination | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, S, Gabelli, S.B, Sohn, J. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The structural basis for 2'-5'/3'-5'-cGAMP synthesis by cGAS
Nat Commun, 15, 2024
|
|
