5ZDF
 
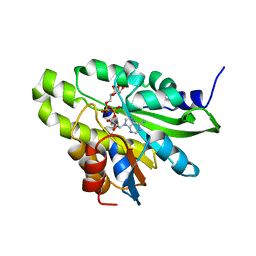 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) T267K mutant from Deinococcus radiodurans in complex with ADP-ribose | | Descriptor: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Cho, C.C, Hsu, C.H. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDS
 
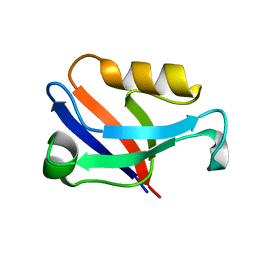 | | Crystal structure of the second PDZ domain of Frmpd2 | | Descriptor: | FERM and PDZ domain-containing 2 | | Authors: | Lu, X, Wang, X.F. | | Deposit date: | 2018-02-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The second PDZ domain of scaffold protein Frmpd2 binds to GluN2A of NMDA receptors.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
8HBA
 
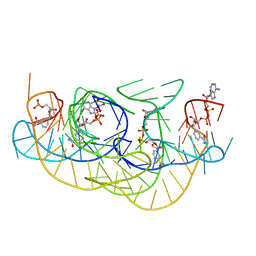 | |
8HIL
 
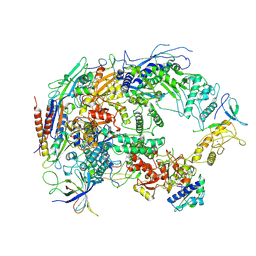 | | A cryo-EM structure of B. oleracea RNA polymerase V at 3.57 Angstrom | | Descriptor: | DNA-dependent RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, DNA-directed RNA polymerase subunit, ... | | Authors: | Du, X, Xie, G, Hu, H, Du, J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
8HB1
 
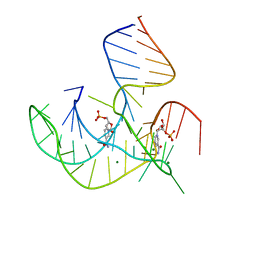 | | Crystal structure of NAD-II riboswitch (two strands) with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, RNA (30-MER), ... | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
8HBD
 
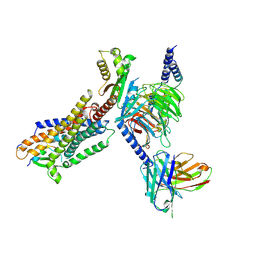 | | Cryo-EM structure of IRL1620-bound ETBR-Gi complex | | Descriptor: | Endothelin receptor type B,Endothelin receptor type B,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Ji, Y, Jiang, Y, Duan, J, Xu, H.E. | | Deposit date: | 2022-10-28 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural basis of peptide recognition and activation of endothelin receptors.
Nat Commun, 14, 2023
|
|
8HJ2
 
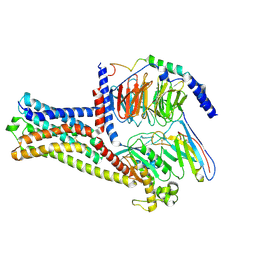 | | GPR21 wt with G15 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X, Xu, F. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
4U5C
 
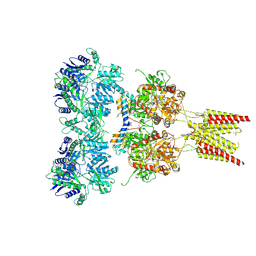 | | Crystal structure of GluA2, con-ikot-ikot snail toxin, partial agonist FW and postitive modulator (R,R)-2b complex | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Con-ikot-ikot, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.6883 Å) | | Cite: | X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism.
Science, 345, 2014
|
|
5ZF9
 
 | | Structure of human dihydroorotate dehydrogenase in complex with 280-12 | | Descriptor: | 3-chloro-4,6-dihydroxy-2-methyl-5-[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]benzaldehyde, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Miyazaki, Y, Inaoka, K.D, Shiba, T, Saimoto, H, Amalia, E, Kido, Y, Sakai, C, Nakamura, M, Moore, L.A, Harada, S, Kita, K. | | Deposit date: | 2018-03-02 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Selective Cytotoxicity of Dihydroorotate Dehydrogenase Inhibitors to Human Cancer Cells Under Hypoxia and Nutrient-Deprived Conditions.
Front Pharmacol, 9, 2018
|
|
8HIX
 
 | | Cryo-EM structure of GPR21_m5_Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X, Xu, F. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
8HB8
 
 | |
8GYX
 
 | | Cryo-EM structure of human CEPT1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Choline/ethanolaminephosphotransferase 1, MAGNESIUM ION | | Authors: | Qian, H.W, Wang, Z.H. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for catalysis of human choline/ethanolamine phosphotransferase 1.
Nat Commun, 14, 2023
|
|
8HCX
 
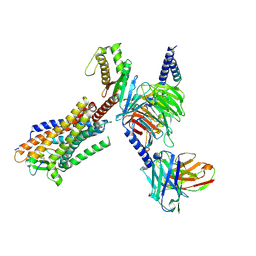 | | Cryo-EM structure of Endothelin1-bound ETBR-Gq complex | | Descriptor: | Endothelin receptor type B,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, Q, Jiang, Y, Xu, H.E, Ji, Y, Duan, J. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of peptide recognition and activation of endothelin receptors.
Nat Commun, 14, 2023
|
|
8HIM
 
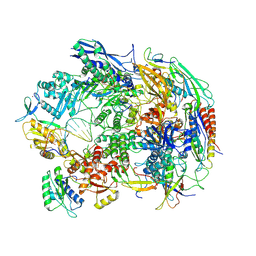 | | A cryo-EM structure of B. oleracea RNA polymerase V elongation complex at 2.73 Angstrom | | Descriptor: | DNA (34-MER), DNA-directed RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, ... | | Authors: | Hu, H, Xie, G, Du, X, Du, J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
8HB3
 
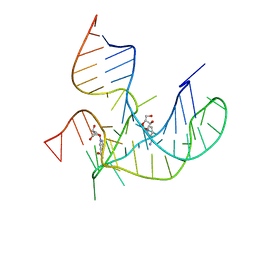 | | Crystal structure of NAD-II riboswitch (two strands) with NR | | Descriptor: | Nicotinamide riboside, RNA (31-MER), RNA (5'-R(*AP*GP*AP*GP*CP*GP*UP*UP*GP*CP*GP*UP*CP*CP*GP*AP*AP*AP*GP*UP*(CBV)P*GP*CP*C)-3') | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
5ZKS
 
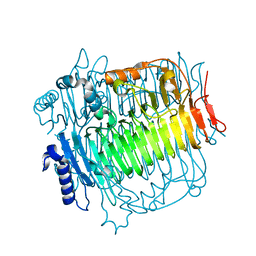 | | Crystal structure of DFA-IIIase from Arthrobacter chlorophenolicus A6 | | Descriptor: | DFA-IIIase | | Authors: | Yu, S.H, Shen, H, Li, X, Mu, W.M. | | Deposit date: | 2018-03-26 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional basis of difructose anhydride III hydrolase, which sequentially converts inulin using the same catalytic residue
Acs Catalysis, 8, 2018
|
|
5ZN3
 
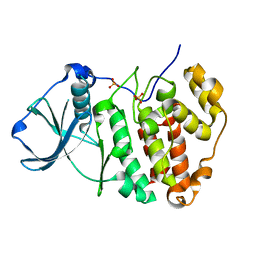 | | X-ray structure of protein kinase ck2 alpha subunit H148S mutant | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN9
 
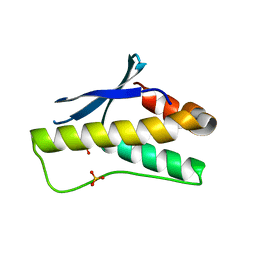 | | Crystal structure of PX domain | | Descriptor: | SULFATE ION, Sorting nexin-27 | | Authors: | Li, Y, Zhu, Z, Li, F, Liao, S, Xu, C. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Crystal structure of PX domain
To Be Published
|
|
7B7F
 
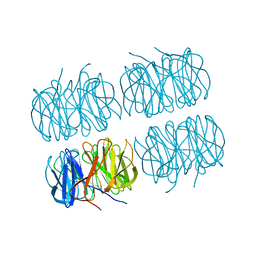 | | Room temperature X-ray structure of H/D-exchanged PLL lectin in complex with L-fucose | | Descriptor: | PLL lectin, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
8HJ0
 
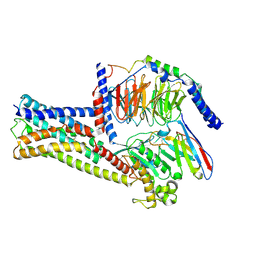 | | GPR21(m5) and G15 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
5HWT
 
 | | Crystal structure of apo-PAS1 | | Descriptor: | Sensor histidine kinase TodS | | Authors: | Hwang, J, Koh, S. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-02 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Insights into Toluene Sensing in the TodS/TodT Signal Transduction System.
J. Biol. Chem., 291, 2016
|
|
8GYW
 
 | | Cryo-EM structure of human CEPT1 complexed with CDP-choline | | Descriptor: | Choline/ethanolaminephosphotransferase 1, MAGNESIUM ION, [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM | | Authors: | Qian, H.W, Wang, Z.H. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for catalysis of human choline/ethanolamine phosphotransferase 1.
Nat Commun, 14, 2023
|
|
5ZNG
 
 | | The crystal complex of immune receptor RGA5A_S of Pia from rice (Oryzae sativa) with rice blast (Magnaporthe oryzae) effector protein AVR1-CO39 | | Descriptor: | AVR1-CO39, NBS-LRR type protein | | Authors: | Guo, L.W, Zhang, Y.K, Liu, Q, Ma, M.Q, Liu, J.F, Peng, Y.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8HCQ
 
 | | Cryo-EM structure of endothelin1-bound ETAR-Gq complex | | Descriptor: | Endothelin-1, Endothelin-1 receptor,Oplophorus-luciferin 2-monooxygenase catalytic subunit chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, Q, Jiang, Y, Xu, H.E, Ji, Y, Duan, J. | | Deposit date: | 2022-11-02 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of peptide recognition and activation of endothelin receptors.
Nat Commun, 14, 2023
|
|
5ZHQ
 
 | |
