2FDG
 
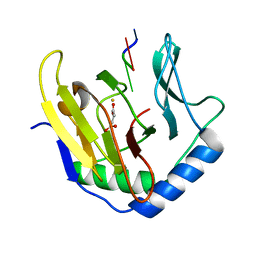 | | Crystal Structure of AlkB in complex with Fe(II), succinate, and methylated trinucleotide T-meA-T | | Descriptor: | 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, FE (II) ION, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2FDI
 
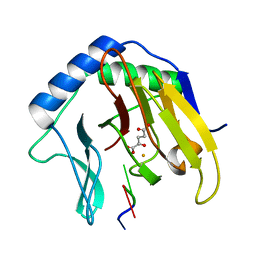 | | Crystal Structure of AlkB in complex with Fe(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T (air 3 hours) | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
4G8K
 
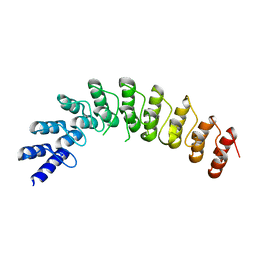 | |
2HB5
 
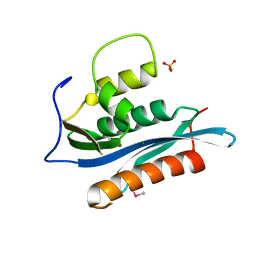 | | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain | | Descriptor: | MAGNESIUM ION, Reverse transcriptase/ribonuclease H, SULFATE ION | | Authors: | Lim, D, Gregorio, G.G, Bingman, C.A, Martinez-Hackert, E, Hendrickson, W.A, Goff, S.P. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of the Moloney Murine Leukemia Virus RNase H Domain.
J.Virol., 80, 2006
|
|
5KS2
 
 | | RAWV_CTD (Helix form) of 16S/23S 2'-O-methyltransferase TlyA | | Descriptor: | 16S/23S rRNA (cytidine-2'-O)-methyltransferase TlyA, CHLORIDE ION | | Authors: | Kuiper, E.G, Conn, G.L. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A Novel Motif for S-Adenosyl-l-methionine Binding by the Ribosomal RNA Methyltransferase TlyA from Mycobacterium tuberculosis.
J. Biol. Chem., 292, 2017
|
|
2XMA
 
 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE RIGHT END DNA COMPLEX | | Descriptor: | DRA2 TRANSPOSASE RIGHT END RECOGNITION SITE, MAGNESIUM ION, TRANSPOSASE | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
5UV5
 
 | | Crystal Structure of a 2-Hydroxyisoquinoline-1,3-dione RNase H Active Site Inhibitor with Multiple Binding Modes to HIV Reverse Transcriptase | | Descriptor: | 7-(furan-2-yl)-2-hydroxyisoquinoline-1,3(2H,4H)-dione, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-02-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A 2-Hydroxyisoquinoline-1,3-Dione Active-Site RNase H Inhibitor Binds in Multiple Modes to HIV-1 Reverse Transcriptase.
Antimicrob. Agents Chemother., 61, 2017
|
|
1XR7
 
 | | Crystal structure of RNA-dependent RNA Polymerase 3D from human rhinovirus serotype 16 | | Descriptor: | Genome polyprotein | | Authors: | Love, R.A, Maegley, K.A, Yu, X, Ferre, R.A, Lingardo, L.K, Diehl, W, Parge, H.E, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2004-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the RNA-Dependent RNA Polymerase from Human Rhinovirus: A Dual-Function Target for Common Cold Antiviral Therapy
Structure, 12, 2004
|
|
2FD8
 
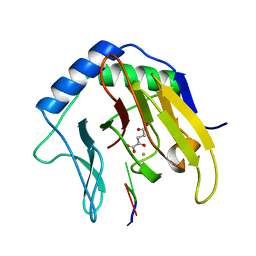 | | Crystal Structure of AlkB in complex with Fe(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
1XR5
 
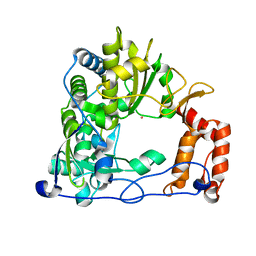 | | Crystal Structure of the RNA-dependent RNA Polymerase 3D from human rhinovirus serotype 14 | | Descriptor: | Genome polyprotein, SAMARIUM (III) ION | | Authors: | Love, R.A, Maegley, K.A, Yu, X, Ferre, R.A, Lingardo, L.K, Diehl, W, Parge, H.E, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2004-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the RNA-Dependent RNA Polymerase from Human Rhinovirus: A Dual-Function Target for Common Cold Antiviral Therapy
Structure, 12, 2004
|
|
1JQS
 
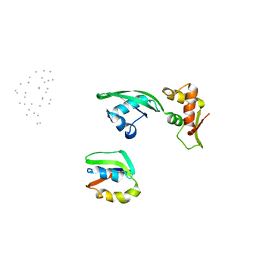 | | Fitting of L11 protein and elongation factor G (domain G' and V) in the cryo-em map of E. coli 70S ribosome bound with EF-G and GMPPCP, a nonhydrolysable GTP analog | | Descriptor: | 50S Ribosomal protein L11, Elongation Factor G | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
5L0Z
 
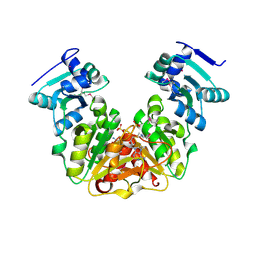 | | Crystal Structure of AdoMet bound rRNA methyltransferase from Sinorhizobium meliloti | | Descriptor: | COBALT (II) ION, Probable RNA methyltransferase, TrmH family, ... | | Authors: | Dey, D, Hegde, R.P, Almo, S.C, Ramakumar, S, Ramagopal, U.A. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of AdoMet bound rRNA methyltransferase from Sinorhizobium meliloti
To Be Published
|
|
2FDH
 
 | | Crystal Structure of AlkB in complex with Mn(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
4CIO
 
 | |
2FDJ
 
 | | Crystal Structure of AlkB in complex with Fe(II) and succinate | | Descriptor: | Alkylated DNA repair protein alkB, FE (II) ION, SUCCINIC ACID | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2FDF
 
 | | Crystal Structure of AlkB in complex with Co(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
2QUD
 
 | | PP7 Coat Protein Dimer | | Descriptor: | Coat protein, GLYCEROL | | Authors: | Chao, J.A. | | Deposit date: | 2007-08-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the coevolution of a viral RNA-protein complex.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2FDK
 
 | | Crystal Structure of AlkB in complex with Fe(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T (air 9 days) | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
4NYN
 
 | |
4ATO
 
 | | New insights into the mechanism of bacterial Type III toxin-antitoxin systems: selective toxin inhibition by a non-coding RNA pseudoknot | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, TOXI, TOXN | | Authors: | Short, F.L, Pei, X.Y, Blower, T.R, Ong, S.L, Luisi, B.F, Salmond, G.P.C. | | Deposit date: | 2012-05-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity and Self-Assembly in the Control of a Bacterial Toxin by an Antitoxic Noncoding RNA Pseudoknot.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4I47
 
 | | Crystal structure of the Ribosome inactivating protein complexed with methylated guanine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-7-methyl-1,7-dihydro-6H-purin-6-one, rRNA N-glycosidase | | Authors: | Yamini, S, Kushwaha, G.S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | First structural evidence of sequestration of mRNA cap structures by type 1 ribosome inactivating protein from Momordica balsamina.
Proteins, 81, 2013
|
|
4BGP
 
 | | Crystal structure of La Crosse virus nucleoprotein | | Descriptor: | GLYCEROL, NUCLEOPROTEIN, SULFATE ION | | Authors: | Reguera, J, Malet, H, Weber, F, Cusack, S. | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-24 | | Last modified: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Encapsidation of Genomic RNA by La Crosse Orthobunyavirus Nucleoprotein
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3ULS
 
 | | Crystal structure of Fab12 | | Descriptor: | Fab12 heavy chain, Fab12 light chain | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
2MJH
 
 | |
9BKD
 
 | | The structure of human Pdcd4 bound to the 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S12, 40S ribosomal protein S13, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Zuber, P, Albacete-Albacete, L, Ramakrishnan, V, S.Fraser, C. | | Deposit date: | 2024-04-27 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Human tumor suppressor protein Pdcd4 binds at the mRNA entry channel in the 40S small ribosomal subunit.
Nat Commun, 15, 2024
|
|
