2B9Z
 
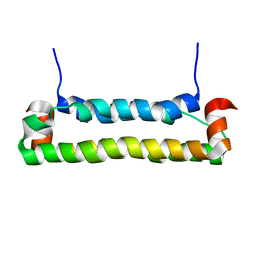 | | Solution structure of FHV B2, a viral suppressor of RNAi | | Descriptor: | B2 protein | | Authors: | Lingel, A, Simon, B, Izaurralde, E, Sattler, M. | | Deposit date: | 2005-10-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition.
Embo Rep., 6, 2005
|
|
2O49
 
 | |
2CQH
 
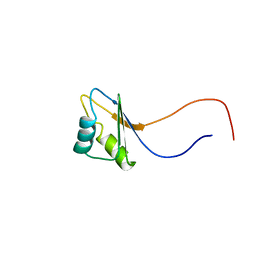 | | Solution structure of the RNA binding domain of IGF-II mRNA-binding protein 2 | | Descriptor: | IGF-II mRNA-binding protein 2 isoform a | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of IGF-II mRNA-binding protein 2
To be Published
|
|
2CQI
 
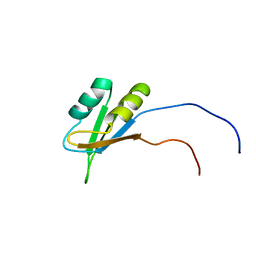 | | Solution structure of the RNA binding domain of Nucleolysin TIAR | | Descriptor: | Nucleolysin TIAR | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain of Nucleolysin TIAR
To be Published
|
|
2DNY
 
 | | Solution structure of the third RNA binding domain of FBP-interacting repressor, SIAHBP1 | | Descriptor: | Fuse-binding protein-interacting repressor, isoform b | | Authors: | Suzuki, S, Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-27 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third RNA binding domain of FBP-interacting repressor, SIAHBP1
To be Published
|
|
2M0Y
 
 | |
2CG7
 
 | | SECOND AND THIRD FIBRONECTIN TYPE I MODULE PAIR (CRYSTAL FORM II). | | Descriptor: | FIBRONECTIN | | Authors: | Rudino-Pinera, E, Ravelli, R.B.G, Sheldrick, G.M, Nanao, M.H, Werner, J.M, Schwarz-Linek, U, Potts, J.R, Garman, E.F. | | Deposit date: | 2006-02-27 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Solution and Crystal Structures of a Module Pair from the Staphylococcus Aureus-Binding Site of Human Fibronectin-A Tale with a Twist.
J.Mol.Biol., 368, 2007
|
|
2CG6
 
 | | Second and third fibronectin type I module pair (crystal form I). | | Descriptor: | HUMAN FIBRONECTIN, SULFATE ION | | Authors: | Rudino-Pinera, E, Ravelli, R.B.G, Sheldrick, G.M, Nanao, M.H, Werner, J.M, Schwarz-Linek, U, Potts, J.R, Garman, E.F. | | Deposit date: | 2006-02-27 | | Release date: | 2007-02-27 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Solution and Crystal Structures of a Module Pair from the Staphylococcus Aureus-Binding Site of Human Fibronectin-A Tale with a Twist.
J.Mol.Biol., 368, 2007
|
|
2L8X
 
 | |
2LK0
 
 | |
2LB9
 
 | | Refined solution structure of a cyanobacterial phytochrome gaf domain in the red light-absorbing ground state (corrected pyrrole ring planarity) | | Descriptor: | PHYCOCYANOBILIN, Sensor histidine kinase | | Authors: | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Vierstra, R.D, Markley, J.L. | | Deposit date: | 2011-03-23 | | Release date: | 2011-06-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cyanobacterial phytochrome GAF domain in the red-light-absorbing ground state.
J.Mol.Biol., 383, 2008
|
|
2LKY
 
 | |
2MFJ
 
 | |
2MB2
 
 | | parallel-stranded G-quadruplex in DNA poly-G stretches | | Descriptor: | DNA_(5'-D(*TP*TP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*T)-3') | | Authors: | Sengar, A, Heddi, B, Phan, A.T. | | Deposit date: | 2013-07-24 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Formation of g-quadruplexes in poly-g sequences: structure of a propeller-type parallel-stranded g-quadruplex formed by a g15 stretch.
Biochemistry, 53, 2014
|
|
2LXS
 
 | | Allosteric communication in the KIX domain proceeds through dynamic re-packing of the hydrophobic core | | Descriptor: | CREB-binding protein, Histone-lysine N-methyltransferase MLL | | Authors: | Bruschweiler, S, Schanda, P, Konrat, R, Tollinger, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core.
Acs Chem.Biol., 8, 2013
|
|
2M6L
 
 | | Solution structure of the Escherichia coli holo ferric enterobactin binding protein | | Descriptor: | Ferrienterobactin-binding periplasmic protein | | Authors: | Chu, B.C.H, Otten, R, Krewulak, K.D, Mulder, F.A.A, Vogel, H.J. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure, binding properties, and dynamics of the bacterial siderophore-binding protein FepB.
J.Biol.Chem., 289, 2014
|
|
2M1C
 
 | | HADDOCK structure of GtYybT PAS Homodimer | | Descriptor: | DHH subfamily 1 protein | | Authors: | Liang, Z.X, Pervushin, K, Tan, E, Rao, F, Pasunooti, S, Soehano, I, Lescar, J. | | Deposit date: | 2012-11-25 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the PAS Domain of a Thermophilic YybT Protein Homolog Reveals a Potential Ligand-binding Site.
J.Biol.Chem., 288, 2013
|
|
2MIW
 
 | | Nuclear magnetic resonance studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2MJ9
 
 | | Designed Exendin-4 analogues | | Descriptor: | Exendin-4 | | Authors: | Rovo, P, Farkas, V, Straner, P, Szabo, M, Jermendy, A, Hegyi, O, Toth, G.K, Perczel, A. | | Deposit date: | 2013-12-30 | | Release date: | 2014-06-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rational design of alpha-helix-stabilized exendin-4 analogues.
Biochemistry, 53, 2014
|
|
2M50
 
 | | Analysis of the structural and molecular basis of voltage-sensitive sodium channel inhibition by the spider toxin, Huwentoxin-IV (-TRTX-Hh2a). | | Descriptor: | Mu-theraphotoxin-Hh2a | | Authors: | Gibbs, A, Minassian, N, Flinspach, M, Wickenden, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | Analysis of the Structural and Molecular Basis of Voltage-sensitive Sodium Channel Inhibition by the Spider Toxin Huwentoxin-IV ( mu-TRTX-Hh2a).
J.Biol.Chem., 288, 2013
|
|
2MEX
 
 | | Structure of the tetrameric building block of the Salmonella Typhimurium PrgI Type three secretion system needle | | Descriptor: | Protein PrgI | | Authors: | Loquet, A, Habenstein, B, Chevelkov, V, Giller, K, Becker, S, Lange, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure and handedness of the building block of a biological assembly.
J.Am.Chem.Soc., 135, 2013
|
|
2LXT
 
 | | Allosteric communication in the KIX domain proceeds through dynamic re-packing of the hydrophobic core | | Descriptor: | CREB-binding protein, Cyclic AMP-responsive element-binding protein 1, Histone-lysine N-methyltransferase MLL | | Authors: | Bruschweiler, S, Schanda, P, Konrat, R, Tollinger, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core.
Acs Chem.Biol., 8, 2013
|
|
2MHJ
 
 | |
2M2D
 
 | | Human programmed cell death 1 receptor | | Descriptor: | Programmed cell death protein 1 | | Authors: | Veverka, V, Cheng, X, Waters, L.C, Muskett, F.W, Morgan, S, Lesley, A, Griffiths, M, Stubberfield, C, Griffin, R, Henry, A.J, Robinson, M.K, Jansson, A, Ladbury, J.E, Ikemizu, S, Davis, S.J, Carr, M.D. | | Deposit date: | 2012-12-18 | | Release date: | 2013-02-27 | | Last modified: | 2013-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the human programmed cell death 1 receptor.
J.Biol.Chem., 288, 2013
|
|
2M3L
 
 | |
