8EHF
 
 | | Cryo-EM structure of his-elemental paused elongation complex with an unfolded TL (1) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6HBI
 
 | | SCAPHARCA DIMERIC HEMOGLOBIN, MUTANT T72V, DEOXY FORM | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Royer Junior, W.E. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational destabilization of the critical interface water cluster in Scapharca dimeric hemoglobin: structural basis for altered allosteric activity.
J.Mol.Biol., 284, 1998
|
|
8EG8
 
 | | Cryo-EM structure of consensus elemental paused elongation complex with a folded TL | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1SJ9
 
 | | Crystal structure of the uridine phosphorylase from Salmonella typhimurium at 2.5A resolution | | Descriptor: | PHOSPHATE ION, Uridine phosphorylase | | Authors: | Dontsova, M, Gabdoulkhakov, A, Morgunova, E, Garber, M, Nikonov, S, Betzel, C, Ealick, S, Mikhailov, A. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preliminary investigation of the three-dimensional structure of Salmonella typhimurium uridine phosphorylase in the crystalline state.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
6HVB
 
 | | NMR structure of Urotensin Peptide Asp-c[Cys-Phe-(N-Me)Trp-Lys-Tyr-Cys]-Val in SDS solution | | Descriptor: | Urotensin-2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Billard, E, Yousif, A.M, Di Maro, S, Abate, L, Bellavita, R, D'Emmanuele di Villa Bianca, R, Santicioli, P, Marinelli, L, Novellino, E, Hebert, T.E, Lubell, W.D, Chatenet, D, Grieco, P. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Functional Selectivity Revealed by N-Methylation Scanning of Human Urotensin II and Related Peptides.
J.Med.Chem., 62, 2019
|
|
6HVK
 
 | | Pepducin UT-Pep2 a biased allosteric agonist of Urotensin-II receptor | | Descriptor: | Urotensin-2 receptor | | Authors: | Carotenuto, A, Hoang, T.A, Nassour, H, Martin, R.D, Billard, E, Myriam, L, Novellino, E, Tanny, J.C, Fournier, A, Hebert, T.E, Chatenet, D. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Lipidated peptides derived from intracellular loops 2 and 3 of the urotensin II receptor act as biased allosteric ligands.
J.Biol.Chem., 297, 2021
|
|
1E8A
 
 | | The three-dimensional structure of human S100A12 | | Descriptor: | CALCIUM ION, S100A12 | | Authors: | Moroz, O.V, Antson, A.A, Murshudov, G.N, Maitland, N.J, Dodson, G.G, Wilson, K.S, Skibshoj, I, Lukanidin, E.M, Bronstein, I.B. | | Deposit date: | 2000-09-18 | | Release date: | 2001-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Three-Dimensional Structure of Human S100A12
Acta Crystallogr.,Sect.D, 57, 2001
|
|
8EHA
 
 | | Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (out) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EGB
 
 | | Cryo-EM structure of consensus elemental paused elongation complex with an unfolded TL | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EG7
 
 | | Cryo-EM structure of pre-consensus elemental paused elongation complex | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-11 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7SEE
 
 | | Structure of E. coli LetB delta (Ring6) mutant, Ring1 in the closed state (Model 1) | | Descriptor: | MCE family protein, Intermembrane transport protein YebT chimera, UNKNOWN ATOM OR ION | | Authors: | Vieni, C, Coudray, N, Bhabha, G, Ekiert, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Role of Ring6 in the Function of the E. coli MCE Protein LetB.
J.Mol.Biol., 434, 2022
|
|
1E5H
 
 | | DELTA-R307A DEACETOXYCEPHALOSPORIN C SYNTHASE COMPLEXED WITH SUCCINATE AND CARBON DIOXIDE | | Descriptor: | CARBON DIOXIDE, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (II) ION, ... | | Authors: | Lee, H.J, Lloyd, M.D, Harlos, K, Clifton, I.J, Baldwin, J.E, Schofield, C.J. | | Deposit date: | 2000-07-26 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinetic and Crystallographic Studies on Deacetoxycephalosporin C Synthase (Daocs)
J.Mol.Biol., 308, 2001
|
|
8EH8
 
 | | Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (1) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EH9
 
 | | Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (2) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7A9A
 
 | | Crystal structure of rubredoxin B (Rv3250c) from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vakhrameev, D, Kavaleuski, A, Bukhdruker, S, Marin, E, Sushko, T, Grabovec, I.P, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2020-09-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A new twist of rubredoxin function in M. tuberculosis.
Bioorg.Chem., 109, 2021
|
|
3N06
 
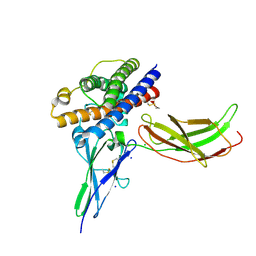 | | A mutant human Prolactin receptor antagonist H27A in complex with the extracellular domain of the human prolactin receptor | | Descriptor: | CHLORIDE ION, Prolactin, Prolactin receptor, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-05-13 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
5ZYF
 
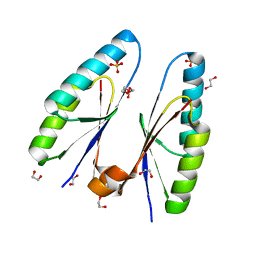 | | Crystal structure of Streptococcus pyogenes type II-A Cas2 | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endoribonuclease Cas2, SULFATE ION | | Authors: | Ka, D, Bae, E. | | Deposit date: | 2018-05-24 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular organization of the type II-A CRISPR adaptation module and its interaction with Cas9 via Csn2
Nucleic Acids Res., 46, 2018
|
|
7Z09
 
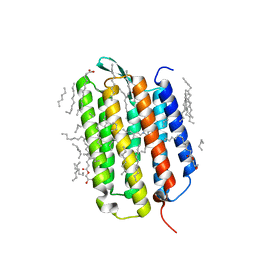 | | Crystal structure of the ground state of bacteriorhodopsin at 1.05 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4KPR
 
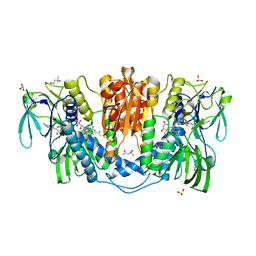 | | Tetrameric form of rat selenoprotein thioredoxin reductase 1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, SULFITE ION, ... | | Authors: | Lindqvist, Y, Sandalova, T, Xu, J, Arner, E. | | Deposit date: | 2013-05-14 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Trp114 residue of thioredoxin reductase 1 is an electron relay sensor for oxidative stress
To be Published, 2013
|
|
1WV0
 
 | | Crystallographic studies on acyl ureas, a new class of inhibitors of glycogen phosphorylase. Broad specificity of the allosteric site | | Descriptor: | 4-[4-({[(2,4-DICHLOROBENZOYL)AMINO]CARBONYL}AMINO)-2,3-DIMETHYLPHENOXY]BUTANOIC ACID, Glycogen phosphorylase, muscle form, ... | | Authors: | Oikonomakos, N.G, Kosmopoulou, M.N, Chrysina, E.D, Leonidas, D.D, Klabunde, T, Wendt, K.U, Defossa, E. | | Deposit date: | 2004-12-10 | | Release date: | 2005-12-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
5W8S
 
 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations | | Descriptor: | LITHIUM ION, Lipid-A-disaccharide synthase, SODIUM ION | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
4XBZ
 
 | | Crystal Structure of EvdO1 from Micromonospora carbonacea var. aurantiaca | | Descriptor: | EvdO1, GLYCEROL, NICKEL (II) ION | | Authors: | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2014-12-17 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7Z0A
 
 | | Crystal structure of the ground state of bacteriorhodopsin at 1.22 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0D
 
 | | Crystal structure of the L state of bacteriorhodopsin at 1.20 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6UC0
 
 | | Isolated S3D-cofilin bound to an actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-09-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
