4LG1
 
 | | Human Methyltransferase-Like Protein 21D | | Descriptor: | 1,2-ETHANEDIOL, Protein-lysine methyltransferase METTL21D, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Zeng, H, Fenner, M, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Human Methyltransferase-Like Protein 21D in Complex with SAM
To be Published
|
|
4FQU
 
 | |
4FPV
 
 | | Crystal structure of D. rerio TDP2 complexed with single strand DNA product | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*G)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4L6H
 
 | |
4HNJ
 
 | |
4KXD
 
 | | Crystal structure of human aminopeptidase A complexed with glutamate and calcium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Y, Liu, C, Lin, Y.Y, Li, F. | | Deposit date: | 2013-05-25 | | Release date: | 2013-07-31 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into central hypertension regulation by human aminopeptidase a.
J.Biol.Chem., 288, 2013
|
|
7BP3
 
 | | Cryo-EM structure of the human MCT2 | | Descriptor: | Monocarboxylate transporter 2 | | Authors: | Zhang, B, Jin, Q, Zhang, X, Guo, J, Ye, S. | | Deposit date: | 2020-03-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cooperative transport mechanism of human monocarboxylate transporter 2.
Nat Commun, 11, 2020
|
|
7BSU
 
 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in PtdSer-bound E2BeF state | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
3IHD
 
 | |
4JKK
 
 | |
6YG8
 
 | |
7BVQ
 
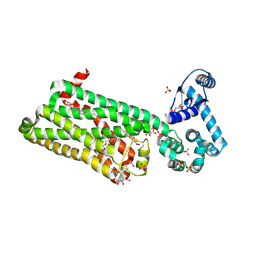 | | Structure of human beta1 adrenergic receptor bound to carazolol | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
6MW3
 
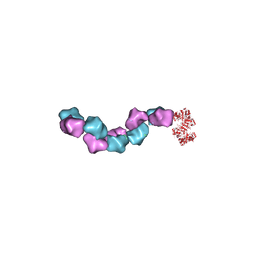 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited filament composed of NrdE alpha subunit and NrdF beta subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase, Ribonucleoside-diphosphate reductase NrdF beta subunit | | Authors: | Thomas, W.C, Bacik, J.P, Kaelber, J.T, Ando, N. | | Deposit date: | 2018-10-29 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
3IK0
 
 | | Lactobacillus casei Thymidylate Synthase in Ternary Complex with dUMP and the Phtalimidic Derivative 7C1 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 4-(4-methyl-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)benzenecarboximidamide, Thymidylate synthase | | Authors: | Pozzi, C, Cancian, L, Leone, R, Luciani, R, Ferrari, S, Mangani, S, Costi, M.P. | | Deposit date: | 2009-08-05 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
7BYM
 
 | | Cryo-EM structure of human KCNQ4 with retigabine | | Descriptor: | Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, POTASSIUM ION, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
3IFB
 
 | | NMR STUDY OF HUMAN INTESTINAL FATTY ACID BINDING PROTEIN | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 1998-10-16 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein: implications for ligand entry and exit.
J.Biomol.NMR, 9, 1997
|
|
7BYS
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
3ILF
 
 | | Crystal structure of porphyranase A (PorA) in complex with neo-porphyrotetraose | | Descriptor: | 6-O-sulfo-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-6-O-sulfo-alpha-L-galactopyranose-(1-3)-alpha-D-galactopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Hehemann, J.H, Correc, G, Barbeyron, T, Helbert, W, Michel, G, Czjzek, M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota.
Nature, 464, 2010
|
|
6XR8
 
 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Volloch, S.R, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
3II0
 
 | | Crystal structure of human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase, cytoplasmic, ... | | Authors: | Ugochukwu, E, Pilka, E, Cooper, C, Bray, J.E, Yue, W.W, Muniz, J, Chaikuad, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-31 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human Glutamate oxaloacetate transaminase 1 (GOT1)
To be Published
|
|
7BZM
 
 | | Crystal structure of rice Os3BGlu7 with glucoimidazole | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, GLUCOIMIDAZOLE, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R, Tankrathok, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Specific Glucoimidazole and Mannoimidazole Binding by Os3BGlu7.
Biomolecules, 10, 2020
|
|
3IJ4
 
 | |
4JQE
 
 | | Crystal structure of scCK2 alpha in complex with AMPPN | | Descriptor: | Casein kinase II subunit alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Liu, H. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HHS
 
 | | Crystal Structure of fatty acid alpha-dioxygenase (Arabidopsis thaliana) | | Descriptor: | Alpha-dioxygenase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Goulah, C.C, Zhu, G, Koszelak-Rosenblum, M, Malkowski, M.G. | | Deposit date: | 2012-10-10 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of alpha-Dioxygenase provides insight into diversity in the cyclooxygenase-peroxidase superfamily.
Biochemistry, 52, 2013
|
|
6XVG
 
 | | Human Sirt6 3-318 in complex with ADP-ribose and the activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, GLYCEROL, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2020-01-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding site for activator MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
