2PL4
 
 | |
3QWG
 
 | |
2MS6
 
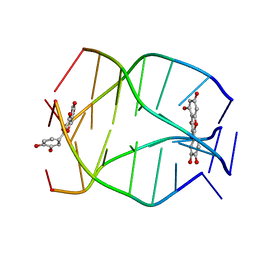 | | Human Telomeric G-quadruplex DNA sequence (TTAGGGT)4 complexed with Flavonoid Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DNA_(5'-D(*TP*TP*AP*GP*GP*GP*T)-3') | | Authors: | Kumar, A, Tawani, A. | | Deposit date: | 2014-07-24 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the interaction of Flavonoids with Human Telomeric Sequence
Sci Rep, 5, 2015
|
|
2EWF
 
 | | Crystal structure of the site-specific DNA nickase N.BspD6I | | Descriptor: | BROMIDE ION, Nicking endonuclease N.BspD6I | | Authors: | Kachalova, G.S, Bartunik, H.D, Artyukh, R.I, Rogulin, E.A, Perevyazova, T.A, Zheleznaya, L.A, Matvienko, N.I. | | Deposit date: | 2005-11-03 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit.
J.Mol.Biol., 384, 2008
|
|
8AXA
 
 | | Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon) | | Descriptor: | Cas12k, DNA non-target strand, DNA target strand, ... | | Authors: | Tenjo-Castano, F, Sofos, N, Stella, S, Fuglsang, A, Pape, T, Mesa, P, Stutzke, L.S, Temperini, P, Montoya, G. | | Deposit date: | 2022-08-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
1ATN
 
 | | Atomic structure of the actin:DNASE I complex | | Descriptor: | ACTIN, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kabsch, W, Mannherz, H.G, Suck, D, Pai, E, Holmes, K.C. | | Deposit date: | 1991-03-08 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of the actin:DNase I complex.
Nature, 347, 1990
|
|
5GVS
 
 | | Crystal structure of the DDX41 DEAD domain in an apo open form | | Descriptor: | Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
5GVR
 
 | | Crystal structure of the DDX41 DEAD domain in an apo closed form | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
2MEM
 
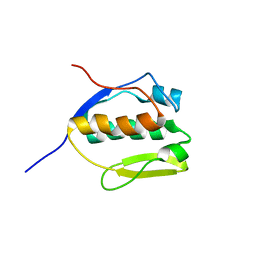 | | Solution NMR structure of SLED domain of Scml2 | | Descriptor: | Sex comb on midleg-like protein 2 | | Authors: | Bezsonova, I. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the DNA-binding Domain from Scml2 (Sex Comb on Midleg-like 2).
J.Biol.Chem., 289, 2014
|
|
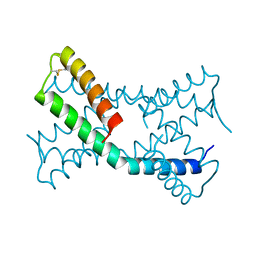
6PSH
 
 | |
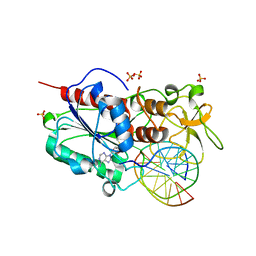
2UYH
 
 | |
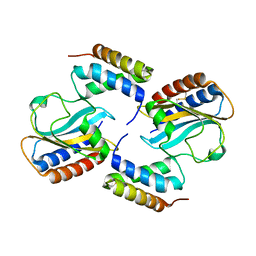
6PXE
 
 | |
6TEH
 
 | | Baseplate of native GTA particle computed with C3 symmetry | | Descriptor: | IRON/SULFUR CLUSTER, Putative gene transfer agent protein | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-12 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TEA
 
 | | Tail of native GTA particle computed with helical refinement, C6 symmetry | | Descriptor: | Phage major tail protein, TP901-1 family | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6PSK
 
 | | Crystal structure of the complex between periplasmic domains of antiholin RI and holin T from T4 phage, in P6522 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Antiholin, ... | | Authors: | Kuznetsov, V.B, Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-06-24 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of T4 Phage Lysis Control: DNA as the Signal for Lysis Inhibition.
J.Mol.Biol., 432, 2020
|
|
6TO8
 
 | | Neck of empty GTA particle computed with C12 symmetry | | Descriptor: | Adaptor protein Rcc01688, Portal protein Rcc01684 | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-12-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TB9
 
 | | Capsid of native GTA particle computed with C5 symmetry | | Descriptor: | Head spike base Rcc01079, Head spike fiber Rcc01080, Major capsid protein Rcc01687 | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6PX4
 
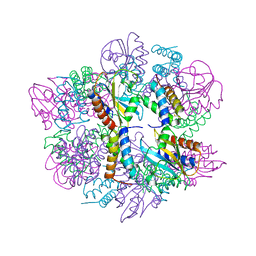 | |
2J63
 
 | | Crystal structure of AP endonuclease LMAP from Leishmania major | | Descriptor: | AP-ENDONUCLEASE | | Authors: | Vidal, A.E, Harkiolaki, M, Gallego, C, Castillo-Acosta, V.M, Ruiz-Perez, L.M, Wilson, K.S, Gonzalez-Pacanowska, D. | | Deposit date: | 2006-09-25 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure and DNA Repair Activities of the Ap Endonuclease from Leishmania Major.
J.Mol.Biol., 373, 2007
|
|
6TSU
 
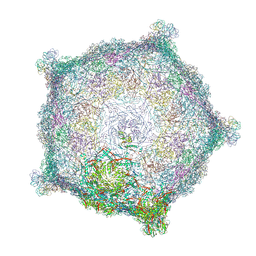 | | Capsid of empty GTA particle computed with C5 symmetry | | Descriptor: | Major capsid protein Rcc01687, Uncharacterized protein | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-12-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
7T5T
 
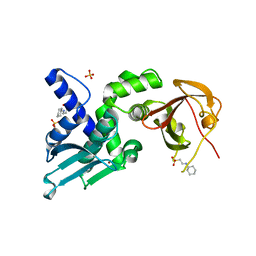 | | Structure of Thauera sp. K11 CapP | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CapP toxin, SULFATE ION, ... | | Authors: | Lau, R.K, Corbett, K.D. | | Deposit date: | 2021-12-13 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A conserved signaling pathway activates bacterial CBASS immune signaling in response to DNA damage.
Embo J., 41, 2022
|
|
2XCO
 
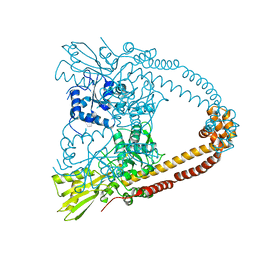 | | The 3.1A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | CALCIUM ION, DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
7Z61
 
 | | Crystal structure of PI3Kgamma with a dihydropurinone inhibitor (compound 18) | | Descriptor: | 9-[(3~{R},4~{R})-4-fluoranylpyrrolidin-3-yl]-7-methyl-2-[(7-methylquinolin-6-yl)amino]purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Goldberg, F.W, Ting, A.K.T, Schimpl, M. | | Deposit date: | 2022-03-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Optimization of hERG and Pharmacokinetic Properties for Basic Dihydro-8 H -purin-8-one Inhibitors of DNA-PK.
Acs Med.Chem.Lett., 13, 2022
|
|
7F9I
 
 | | The apo-form structure of EnrR | | Descriptor: | EnrR repressor | | Authors: | Gan, J.H, Wang, Q.Y. | | Deposit date: | 2021-07-04 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Xenogeneic nucleoid-associated EnrR thwarts H-NS silencing of bacterial virulence with unique DNA binding.
Nucleic Acids Res., 50, 2022
|
|
2VLD
 
 | | crystal structure of a repair endonuclease from Pyrococcus abyssi | | Descriptor: | Endonuclease NucS | | Authors: | Ren, B, Kuhn, J, Meslet-Cladiere, L, Briffotaux, J, Norais, C, Lavigne, R, Flament, D, Ladenstein, R, Myllykallio, H. | | Deposit date: | 2008-01-14 | | Release date: | 2009-05-19 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of a novel endonuclease acting on branched DNA substrates.
EMBO J., 28, 2009
|
|
