3T9D
 
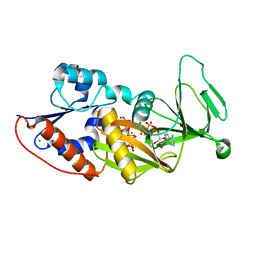 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP and 5-(PP)-IP5 (5-IP7) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
7QTT
 
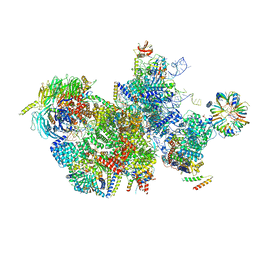 | |
3T9A
 
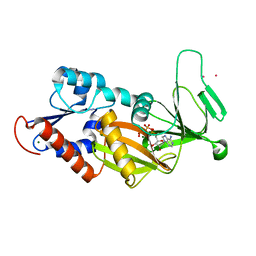 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP at pH 7.0 | | Descriptor: | CADMIUM ION, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
7V66
 
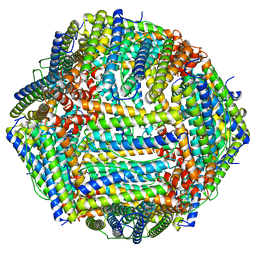 | | Structure of Apoferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, X, Wu, C, Shi, H. | | Deposit date: | 2021-08-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Low-cooling-rate freezing in biomolecular cryo-electron microscopy for recovery of initial frames.
QRB Discov, 2, 2021
|
|
7PFP
 
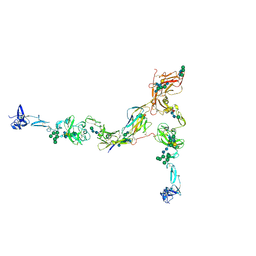 | | Full-length cryo-EM structure of the native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jovine, L, Xu, C, Stsiapanava, A, Carroni, M, Tunyasuvunakool, K, Jumper, J, Wu, B. | | Deposit date: | 2021-08-11 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7T6V
 
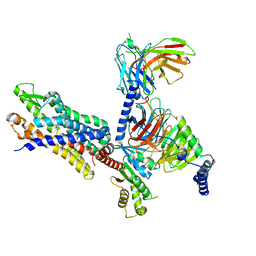 | | Structure of the human FPR2-Gi complex with fMLFII | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7T6U
 
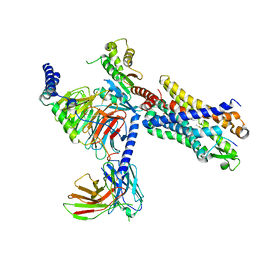 | | Structure of the human FPR2-Gi complex with CGEN-855A | | Descriptor: | B9-scFv, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7T6T
 
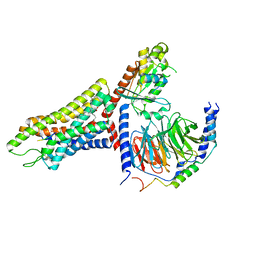 | | Structure of the human FPR1-Gi complex with fMLFII | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
7SZZ
 
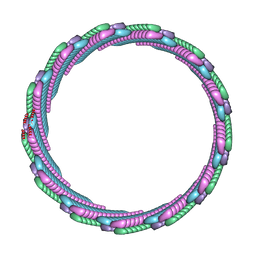 | | Structure of the smaller diameter PSMalpha3 nanotubes | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Beltran, L.C, Kreutzberger, M.A, Wang, S, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-11-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Phenol-soluble modulins PSM alpha 3 and PSM beta 2 form nanotubes that are cross-alpha amyloids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7T8U
 
 | |
7T0X
 
 | | Structure of the larger diameter PSMalpha3 nanotube | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Kreutzberger, M.A, Wang, S, Beltran, L.C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2021-11-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Phenol-soluble modulins PSM alpha 3 and PSM beta 2 form nanotubes that are cross-alpha amyloids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TYV
 
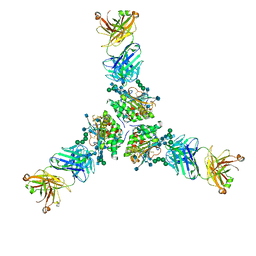 | | Structure of Lassa Virus glycoprotein (Josiah) bound to Fab 25.10C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 25.10C Fab Heavy Chain, ... | | Authors: | Enriquez, A.S, Hastie, K.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-06-01 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Delineating the mechanism of anti-Lassa virus GPC-A neutralizing antibodies.
Cell Rep, 39, 2022
|
|
8WZH
 
 | |
7W0M
 
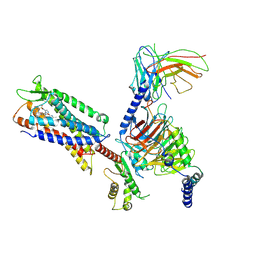 | | Cryo-EM structure of a monomeric GPCR-Gi complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Wu, L.J, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7T20
 
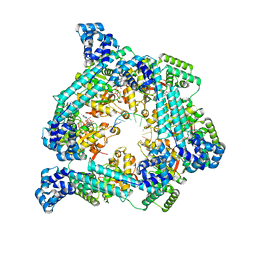 | | E. coli DnaB bound to ssDNA and AMPPNP | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Oakley, A.J, Xu, Z.Q. | | Deposit date: | 2021-12-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Sequence of conformation changes of DnaB helicase during DNA unwinding and priming in Escherichia coli
To Be Published
|
|
7T6S
 
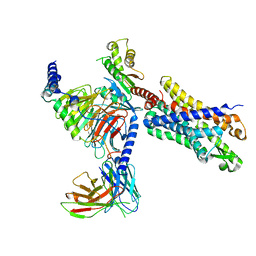 | | Structure of the human FPR2-Gi complex with compound C43 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y.W. | | Deposit date: | 2021-12-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular recognition of formylpeptides and diverse agonists by the formylpeptide receptors FPR1 and FPR2.
Nat Commun, 13, 2022
|
|
4W8F
 
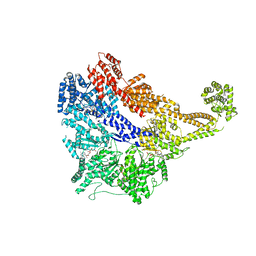 | | Crystal structure of the dynein motor domain in the AMPPNP-bound state | | Descriptor: | Dynein heavy chain lysozyme chimera, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Cheng, H.-C, Bhabha, G, Zhang, N, Vale, R.D. | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.541 Å) | | Cite: | Allosteric communication in the Dynein motor domain.
Cell, 159, 2014
|
|
6SIC
 
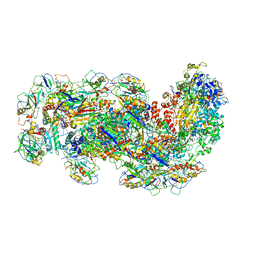 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr1 family, Cmr4 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
8SR5
 
 | |
5SJU
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 6-cyclopropyl-N-[4-(piperidin-1-ylmethyl)-1,3-thiazol-2-yl]-3-(pyrimidin-5-ylamino)pyrazine-2-carboxamide | | Descriptor: | 6-cyclopropyl-N-{(4S)-4-[(piperidin-1-yl)methyl]-4,5-dihydro-1,3-thiazol-2-yl}-3-[(pyrimidin-5-yl)amino]pyrazine-2-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SFQ
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 4-(azetidine-1-carbonyl)-N-[1-(2,2-difluoroethyl)pyrazol-3-yl]-2-methylpyrazole-3-carboxamide | | Descriptor: | 4-(azetidine-1-carbonyl)-N-[1-(2,2-difluoroethyl)-1H-pyrazol-3-yl]-1-methyl-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SKB
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 9-methoxy-3,5-dihydropyrimido[5,4-b]indol-4-one | | Descriptor: | 9-methoxy-3,5-dihydro-4H-pyrimido[5,4-b]indol-4-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Brunner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SG8
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 5-butoxy-4-ethoxypyrimidin-2-amine | | Descriptor: | 5-butoxy-4-ethoxypyrimidin-2-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Gruessner, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SGP
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 4-N-(2-methoxyethyl)-4-N,2-dimethyl-3-N-(2-phenylimidazo[1,2-a]pyridin-7-yl)pyrazole-3,4-dicarboxamide | | Descriptor: | GLYCEROL, MAGNESIUM ION, N~4~-(2-methoxyethyl)-N~4~,1-dimethyl-N~5~-[(4R)-2-phenylimidazo[1,2-a]pyridin-7-yl]-1H-pyrazole-4,5-dicarboxamide, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Peters, J, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SKR
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 1-methyl-N-(2-methyl-5-pyridin-2-ylpyrazol-3-yl)-4-pyridin-4-ylpyrazole-3-carboxamide | | Descriptor: | 1-methyl-N-[1-methyl-3-(pyridin-2-yl)-1H-pyrazol-5-yl]-4-(pyridin-4-yl)-1H-pyrazole-3-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Koerner, M, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
