2Y9M
 
 | | Pex4p-Pex22p structure | | Descriptor: | 1,2-ETHANEDIOL, PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-10-26 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into Ubiquitin-Conjugating Enzyme/ Co-Activator Interactions from the Structure of the Pex4P:Pex22P Complex.
Embo J., 31, 2011
|
|
2YGN
 
 | | WIF domain of human Wnt inhibitory factor 1 in complex with 1,2- dipalmitoylphosphatidylcholine | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Malinauskas, T, Aricescu, A.R, Lu, W, Siebold, C, Jones, E.Y. | | Deposit date: | 2011-04-19 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Modular Mechanism of Wnt Signaling Inhibition by Wnt Inhibitory Factor 1
Nat.Struct.Mol.Biol., 18, 2011
|
|
1BG2
 
 | | HUMAN UBIQUITOUS KINESIN MOTOR DOMAIN | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, KINESIN, ... | | Authors: | Kull, F.J, Sablin, E.P, Lau, R, Fletterick, R.J, Vale, R.D. | | Deposit date: | 1998-06-04 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the kinesin motor domain reveals a structural similarity to myosin.
Nature, 380, 1996
|
|
2DSB
 
 | |
2YV6
 
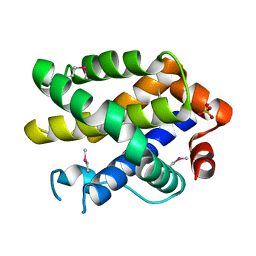 | | Crystal structure of human Bcl-2 family protein Bak | | Descriptor: | Bcl-2 homologous antagonist/killer, SULFATE ION | | Authors: | Wang, H, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-09 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel dimerization mode of the human Bcl-2 family protein Bak, a mitochondrial apoptosis regulator.
J.Struct.Biol., 166, 2009
|
|
2YXO
 
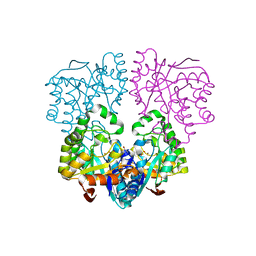 | | Histidinol Phosphate Phosphatase complexed with Sulfate | | Descriptor: | FE (III) ION, GLYCEROL, Histidinol phosphatase, ... | | Authors: | Omi, R. | | Deposit date: | 2007-04-26 | | Release date: | 2007-11-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Monofunctional Histidinol Phosphate Phosphatase from Thermus thermophilus HB8.
Biochemistry, 46, 2007
|
|
2YZ5
 
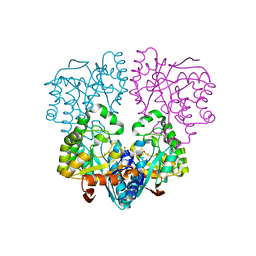 | | Histidinol Phosphate Phosphatase complexed with Phosphate | | Descriptor: | FE (III) ION, GLYCEROL, Histidinol phosphatase, ... | | Authors: | Omi, R. | | Deposit date: | 2007-05-03 | | Release date: | 2007-11-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Monofunctional Histidinol Phosphate Phosphatase from Thermus thermophilus HB8.
Biochemistry, 46, 2007
|
|
2YLE
 
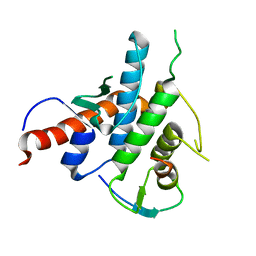 | | Crystal structure of the human Spir-1 KIND FSI domain in complex with the FSI peptide | | Descriptor: | FORMIN-2, PROTEIN SPIRE HOMOLOG 1 | | Authors: | Zeth, K, Pechlivanis, M, Vonrhein, C, Kerkhoff, E. | | Deposit date: | 2011-06-01 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Actin Nucleation Factor Cooperativity: Crystal Structure of the Spir-1 Kinase Non-Catalytic C-Lobe Domain (Kind)Formin-2 Formin Spir Interaction Motif (Fsi) Complex.
J.Biol.Chem., 286, 2011
|
|
2ZXE
 
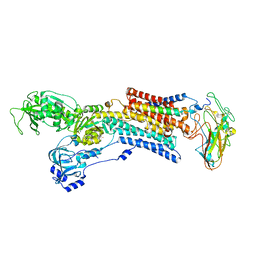 | | Crystal structure of the sodium - potassium pump in the E2.2K+.Pi state | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Shinoda, T, Ogawa, H, Cornelius, F, Toyoshima, C. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the sodium - potassium pump at 2.4 A resolution
Nature, 459, 2009
|
|
4Q4E
 
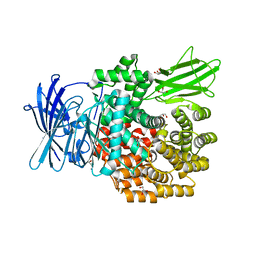 | | Crystal structure of E.coli aminopeptidase N in complex with actinonin | | Descriptor: | ACTINONIN, Aminopeptidase N, GLYCEROL, ... | | Authors: | Reddi, R, Ganji, R.J, Addlagatta, A. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of M1 family aminopeptidases by the natural product actinonin: Crystal structure in complex with E. coli aminopeptidase N.
Protein Sci., 24, 2015
|
|
2YXW
 
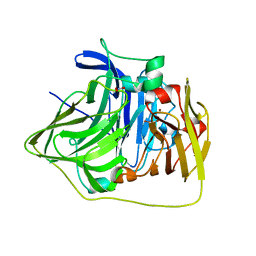 | | The deletion mutant of Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Higuchi, Y, Komori, H. | | Deposit date: | 2007-04-27 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site
J.Mol.Biol., 373, 2007
|
|
2YHF
 
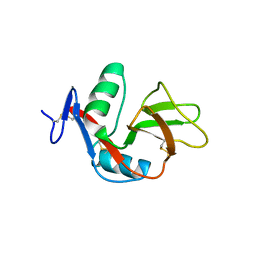 | | 1.9 Angstrom Crystal Structure of CLEC5A | | Descriptor: | C-TYPE LECTIN DOMAIN FAMILY 5 MEMBER A | | Authors: | Watson, A.A, Lebedev, A.A, Murshudov, G.M, Vagin, A.A, Hall, B.A, O'Callaghan, C.A. | | Deposit date: | 2011-04-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Flexibility of the Macrophage Dengue Virus Receptor Clec5A: Implications for Ligand Binding and Signaling.
J.Biol.Chem., 286, 2011
|
|
2DDR
 
 | | Crystal structure of sphingomyelinase from Bacillus cereus with calcium ion | | Descriptor: | CALCIUM ION, Sphingomyelin phosphodiesterase | | Authors: | Ago, H, Oda, M, Takahashi, M, Tsuge, H, Ochi, S, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-02 | | Release date: | 2006-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of the Sphingomyelin Phosphodiesterase Activity in Neutral Sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
2DG5
 
 | | Crystal Structure of Gamma-glutamyl transpeptidase from Escherichia coli in complex with hydrolyzed Glutathione | | Descriptor: | CALCIUM ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Okada, T, Wada, K, Fukuyama, K. | | Deposit date: | 2006-03-08 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1AUO
 
 | | CARBOXYLESTERASE FROM PSEUDOMONAS FLUORESCENS | | Descriptor: | CARBOXYLESTERASE | | Authors: | Kim, K.K, Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-01 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of carboxylesterase from Pseudomonas fluorescens, an alpha/beta hydrolase with broad substrate specificity.
Structure, 5, 1997
|
|
3ICT
 
 | | Crystal structure of reduced Bacillus anthracis CoADR-RHD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, Coenzyme A-Disulfide Reductase, ... | | Authors: | Wallen, J.R, Claiborne, A. | | Deposit date: | 2009-07-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking.
Biochemistry, 48, 2009
|
|
2ZXP
 
 | | Crystal structure of RecJ in complex with Mn2+ from Thermus thermophilus HB8 | | Descriptor: | MANGANESE (II) ION, Single-stranded DNA specific exonuclease RecJ | | Authors: | Wakamatsu, T, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of RecJ exonuclease defines its specificity for single-stranded DNA
J.Biol.Chem., 285, 2010
|
|
2DPT
 
 | |
4QNC
 
 | | Crystal structure of a SemiSWEET in an occluded state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PENTADECANE, chemical transport protein | | Authors: | Yan, X, Yuyong, T, Liang, F, Perry, K. | | Deposit date: | 2014-06-17 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structures of bacterial homologues of SWEET transporters in two distinct conformations.
Nature, 515, 2014
|
|
1WOM
 
 | | Crystal structure of RsbQ | | Descriptor: | MALONIC ACID, S-1,2-PROPANEDIOL, Sigma factor sigB regulation protein rsbQ | | Authors: | Kaneko, T, Kumasaka, T, Tanaka, N. | | Deposit date: | 2004-08-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis
Protein Sci., 14, 2005
|
|
2DSC
 
 | |
1ITG
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HIV-1 INTEGRASE: SIMILARITY TO OTHER POLYNUCLEOTIDYL TRANSFERASES | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Dyda, F, Hickman, A.B, Jenkins, T.M, Engelman, A, Craigie, R, Davies, D.R. | | Deposit date: | 1994-11-21 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases.
Science, 266, 1994
|
|
4Q4I
 
 | | Crystal structure of E.coli aminopeptidase N in complex with amastatin | | Descriptor: | Amastatin, Aminopeptidase N, GLYCEROL, ... | | Authors: | Reddi, R, Ganji, R.J, Addlagatta, A. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the inhibition of M1 family aminopeptidases by the natural product actinonin: Crystal structure in complex with E. coli aminopeptidase N.
Protein Sci., 24, 2015
|
|
1BQU
 
 | | CYTOKYNE-BINDING REGION OF GP130 | | Descriptor: | GLYCEROL, PROTEIN (GP130), SULFATE ION | | Authors: | Bravo, J, Staunton, D, Heath, J.K, Jones, E.Y. | | Deposit date: | 1998-08-18 | | Release date: | 1998-08-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cytokine-binding region of gp130.
EMBO J., 17, 1998
|
|
2DPS
 
 | |
