5TBB
 
 | |
5TBA
 
 | |
5TB8
 
 | |
5TBC
 
 | |
9EZZ
 
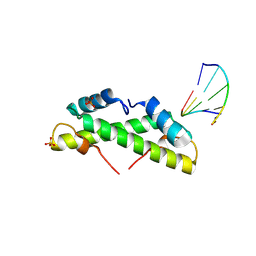 | | Bacterial histone protein HBb from Bdellovibrio bacteriovorus bound to DNA | | Descriptor: | DNA (5'-D(P*AP*GP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*CP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hu, Y, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2024-04-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bacterial histone HBb from Bdellovibrio bacteriovorus compacts DNA by bending.
Nucleic Acids Res., 52, 2024
|
|
9F0E
 
 | |
4WUL
 
 | |
4WU4
 
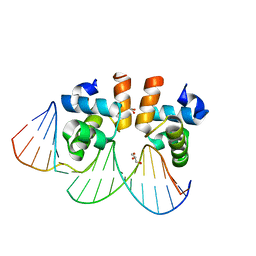 | | Crystal structure of E. faecalis DNA binding domain LiaRD191N complexed with 22bp DNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*CP*GP*TP*TP*CP*TP*TP*AP*AP*GP*TP*CP*C)-3'), DNA (5'-D(P*GP*GP*AP*CP*TP*TP*AP*AP*GP*AP*AP*CP*GP*AP*TP*TP*T)-3'), GLYCEROL, ... | | Authors: | Davlieva, M, Shamoo, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin.
Nucleic Acids Res., 43, 2015
|
|
7ICF
 
 | |
7ICJ
 
 | |
7ICU
 
 | |
1SKN
 
 | | THE BINDING DOMAIN OF SKN-1 IN COMPLEX WITH DNA: A NEW DNA-BINDING MOTIF | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*GP*AP*TP*GP*AP*CP*AP*TP*TP*GP*T)-3'), DNA (5'-D(*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*TP*CP*CP*C)-3'), DNA-BINDING DOMAIN OF SKN-1, ... | | Authors: | Rupert, P.B, Daughdrill, G.W, Bowerman, B, Matthews, B.W. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new DNA-binding motif in the Skn-1 binding domain-DNA complex.
Nat.Struct.Biol., 5, 1998
|
|
6T8H
 
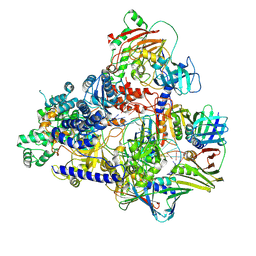 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
2IIE
 
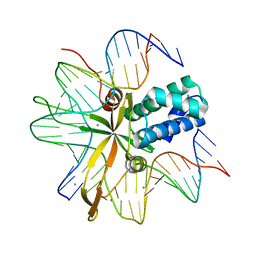 | | single chain Integration Host Factor protein (scIHF2) in complex with DNA | | Descriptor: | DNA (5'-D(*DGP*DCP*DTP*DTP*DAP*DTP*DCP*DAP*DAP*DTP*DTP*DTP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DCP*DAP*DAP*DAP*DAP*DAP*DAP*DGP*DCP*DAP*DTP*DT)-3'), Integration host factor, ... | | Authors: | Bao, Q, Droege, P, Davey, C.A. | | Deposit date: | 2006-09-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A Divalent Metal-mediated Switch Controlling Protein-induced DNA Bending
J.Mol.Biol., 367, 2007
|
|
2IIF
 
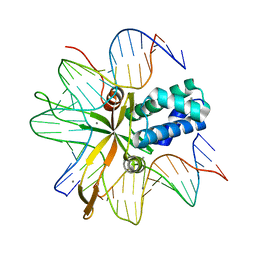 | | single chain Integration Host Factor mutant protein (scIHF2-K45aE) in complex with DNA | | Descriptor: | DNA (5'-D(*DGP*DCP*DTP*DTP*DAP*DTP*DCP*DAP*DAP*DTP*DTP*DTP*DGP*DTP*DTP*DGP*DCP*DAP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DCP*DAP*DAP*DAP*DAP*DAP*DAP*DGP*DCP*DAP*DTP*DT)-3'), Integration host factor, ... | | Authors: | Bao, Q, Droege, P, Davey, C.A. | | Deposit date: | 2006-09-28 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Divalent Metal-mediated Switch Controlling Protein-induced DNA Bending
J.Mol.Biol., 367, 2007
|
|
4QTR
 
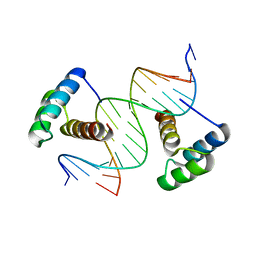 | | Computational design of co-assembling protein-DNA nanowires | | Descriptor: | DNA (5'-D(P*CP*GP*GP*AP*AP*AP*TP*TP*AP*AP*AP*TP*TP*AP*CP*A)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*AP*TP*TP*TP*AP*AP*TP*TP*TP*CP*C)-3'), dualENH | | Authors: | Mou, Y, Mayo, S.L. | | Deposit date: | 2014-07-08 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Computational design of co-assembling protein-DNA nanowires.
Nature, 525, 2015
|
|
7PBK
 
 | | Vibriophage phiVC8 family A DNA polymerase (DpoZ), two conformations: thumb-exo open and thumb-exo closed | | Descriptor: | DNA polymerase I | | Authors: | Czernecki, D, Hu, H, Romoli, F, Delarue, M. | | Deposit date: | 2021-08-02 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage phi VC8.
Nucleic Acids Res., 49, 2021
|
|
1HCQ
 
 | | THE CRYSTAL STRUCTURE OF THE ESTROGEN RECEPTOR DNA-BINDING DOMAIN BOUND TO DNA: HOW RECEPTORS DISCRIMINATE BETWEEN THEIR RESPONSE ELEMENTS | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*TP*CP*AP*CP*AP*GP*TP*GP*AP*CP*CP*T P*G)-3'), DNA (5'-D(*CP*CP*AP*GP*GP*TP*CP*AP*CP*TP*GP*TP*GP*AP*CP*CP*T P*G)-3'), PROTEIN (ESTROGEN RECEPTOR), ... | | Authors: | Schwabe, J.W.R, Chapman, L, Finch, J.T, Rhodes, D. | | Deposit date: | 1995-01-04 | | Release date: | 1995-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements.
Cell(Cambridge,Mass.), 75, 1993
|
|
4TUP
 
 | |
1OCT
 
 | | CRYSTAL STRUCTURE OF THE OCT-1 POU DOMAIN BOUND TO AN OCTAMER SITE: DNA RECOGNITION WITH TETHERED DNA-BINDING MODULES | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*TP*AP*TP*TP*TP*GP*CP*AP*TP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*CP*AP*AP*AP*TP*AP*AP*GP*G)-3'), PROTEIN (OCT-1 POU DOMAIN) | | Authors: | Klemm, J.D, Rould, M.A, Aurora, R, Herr, W, Pabo, C.O. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules.
Cell(Cambridge,Mass.), 77, 1994
|
|
8V6G
 
 | | DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.16 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6H
 
 | | DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7OPL
 
 | |
6N2S
 
 | |
1LBG
 
 | | LACTOSE OPERON REPRESSOR BOUND TO 21-BASE PAIR SYMMETRIC OPERATOR DNA, ALPHA CARBONS ONLY | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), PROTEIN (LACTOSE OPERON REPRESSOR) | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-01-03 | | Release date: | 1996-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
