3J4Q
 
 | |
3VD6
 
 | | Both Zn Fingers of GATA1 Bound to Palindromic DNA Recognition Site, P21 Crystal Form | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*AP*GP*AP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*TP*C)-3'), ... | | Authors: | Jacques, D.A, Ripin, N, Wilkinson-White, L.E, Guss, J.M, Matthews, J.M. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Protein Sci., 24, 2015
|
|
7M2E
 
 | | Crystal structure of BPTF bromodomain in complex with CB02-092 | | Descriptor: | 4-chloro-5-{4-[2-(dimethylamino)ethyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Nithianantham, S, Fischer, M. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition
J.Med.Chem., 64, 2021
|
|
3VEK
 
 | | Both Zn Fingers of GATA1 Bound to Palindromic DNA Recognition Site, P1 Crystal Form | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*TP*C)-3'), Erythroid transcription factor, ... | | Authors: | Jacques, D.A, Ripin, N, Wilkinson-White, L.E, Guss, J.M, Matthews, J.M. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Protein Sci., 24, 2015
|
|
3ZMV
 
 | | LSD1-CoREST in complex with PLSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PKSFLV PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
5HLN
 
 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH CHIR99021 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHIR99021, Glycogen synthase kinase-3 beta, ... | | Authors: | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-25 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
3ZN1
 
 | | LSD1-CoREST in complex with PRLYLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3FGH
 
 | | Human mitochondrial transcription factor A box B | | Descriptor: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gangelhoff, T.A, Mungalachetty, P, Nix, J, Churchill, M.E.A. | | Deposit date: | 2008-12-06 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A
Nucleic Acids Res., 37, 2009
|
|
3J4R
 
 | |
2O8M
 
 | | Crystal structure of the S139A mutant of Hepatitis C Virus NS3/4A protease | | Descriptor: | Protease, SODIUM ION, ZINC ION | | Authors: | Fischmann, T.O, Prongay, A.J, Madison, V.M, Yao, N. | | Deposit date: | 2006-12-12 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization
J.Med.Chem., 50, 2007
|
|
2MTS
 
 | | Three-Dimensional Structure and Interaction Studies of Hepatitis C Virus p7 in 1,2-Dihexanoyl-sn-glycero-3-phosphocholine by Solution Nuclear Magnetic Resonance | | Descriptor: | HEPATITIS C VIRUS P7 PROTEIN | | Authors: | Cook, G.A, Dawson, L.A, Tian, Y, Opella, S.J. | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and interaction studies of hepatitis C virus p7 in 1,2-dihexanoyl-sn-glycero-3-phosphocholine by solution nuclear magnetic resonance.
Biochemistry, 52, 2013
|
|
3ZN0
 
 | | LSD1-CoREST in complex with PRSFAA peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZP5
 
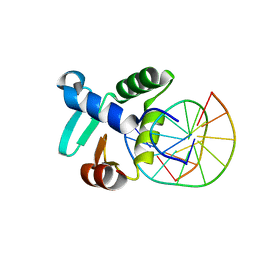 | | Crystal structure of the DNA binding ETS domain of the human protein FEV in complex with DNA | | Descriptor: | 5'-D(*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP)-3', 5'-D(*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP)-3', PROTEIN FEV | | Authors: | Newman, J.A, Cooper, C.D.O, Krojer, T, Shrestha, L, Allerston, C.K, Vollmar, M, Arrowsmith, C.H, Burgess-Brown, N, Edwards, A, von Delft, F, Gileadi, O. | | Deposit date: | 2013-02-26 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
2N1P
 
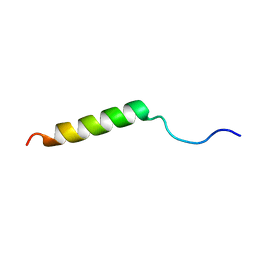 | |
3ZMU
 
 | | LSD1-CoREST in complex with PKSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PKSFLV PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZMZ
 
 | | LSD1-CoREST in complex with PRSFAV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
2R6M
 
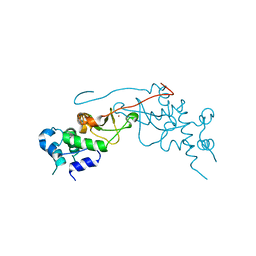 | | Crystal structure of rat CK2-beta subunit | | Descriptor: | Casein kinase II subunit beta, ZINC ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
3UX0
 
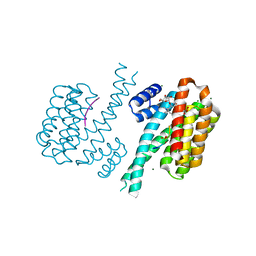 | | Crystal structure of human 14-3-3 sigma in complex with TASK-3 peptide and stabilizer Fusicoccin H | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(hydroxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-gulopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Bartel, M, Anders, C, Higuchi, Y, Schumacher, B, Kato, N, Ottmann, C. | | Deposit date: | 2011-12-03 | | Release date: | 2013-01-02 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of k(+) channels at the cell surface.
Chem.Biol., 20, 2013
|
|
1SVB
 
 | |
3V61
 
 | | Structure of S. cerevisiae PCNA conjugated to SUMO on lysine 164 | | Descriptor: | BARIUM ION, N-ETHYLMALEIMIDE, Proliferating cell nuclear antigen, ... | | Authors: | Armstrong, A.A, Mohideen, F, Lima, C.D. | | Deposit date: | 2011-12-18 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2.
Nature, 483, 2012
|
|
3ZMT
 
 | | LSD1-CoREST in complex with PRSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
2K8J
 
 | | Solution structure of HCV p7 tm2 | | Descriptor: | p7tm2 | | Authors: | Montserret, R, Penin, F. | | Deposit date: | 2008-09-12 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and ion channel activity of the p7 protein from hepatitis C virus.
J.Biol.Chem., 285, 2010
|
|
3O6Q
 
 | | The Structure of SpoIISA and SpoIISB, a Toxin - Antitoxin System | | Descriptor: | Stage II sporulation protein SA, Stage II sporulation protein SB | | Authors: | Levdikov, V.M, Blagova, E.V, Lebedev, A.A, Wilkinson, A.J, Florek, P, Barak, I. | | Deposit date: | 2010-07-29 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure and interactions of SpoIISA and SpoIISB, a toxin-antitoxin system in Bacillus subtilis.
J.Biol.Chem., 161, 2010
|
|
4AP4
 
 | | Rnf4 - ubch5a - ubiquitin heterotrimeric complex | | Descriptor: | E3 UBIQUITIN LIGASE RNF4, UBIQUITIN C, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Plechanovova, A, Hay, R.T, Tatham, M.H, Jaffray, E, Naismith, J.H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of a Ring E3 Ligase and Ubiquitin-Loaded E2 Primed for Catalysis
Nature, 489, 2012
|
|
2M6X
 
 | |
