7Y5E
 
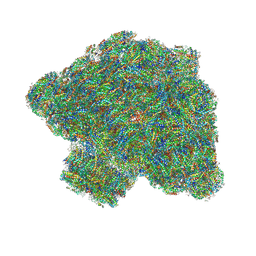 | | In situ single-PBS-PSII-PSI-LHCs megacomplex. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sui, S.F. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7XXO
 
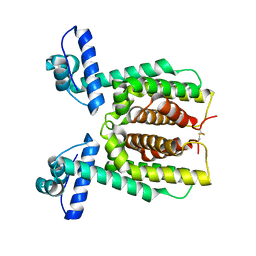 | | HapR Native in CHES buffer pH 9.5 | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XY5
 
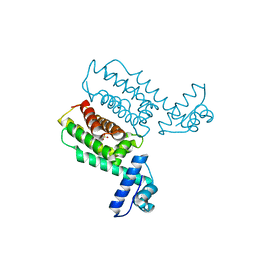 | | HapR_Double Mutant with CHES buffer | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXT
 
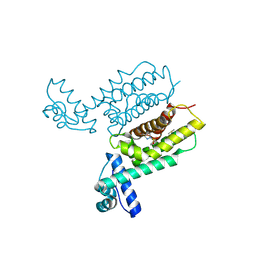 | | HapR Quadruple mutant, bound to IMTVC-212 | | Descriptor: | 1-((5-phenylthiophen-2-yl)sulfonyl)-1H-pyrazole, DIMETHYL SULFOXIDE, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XW3
 
 | |
5KLB
 
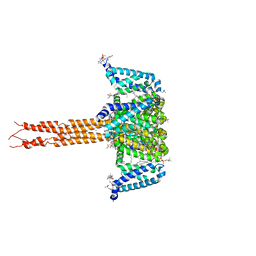 | | Crystal structure of the CavAb voltage-gated calcium channel(wild-type, 2.7A) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CALCIUM ION, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
7XW2
 
 | |
2C5F
 
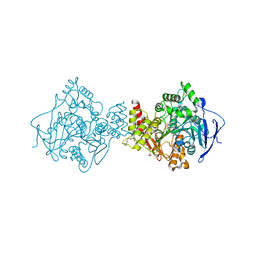 | | Torpedo californica acetylcholinesterase in complex with a non hydrolysable substrate analogue, 4-oxo-N,N,N-trimethylpentanaminium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Fournier, D, Greenblatt, H.M, Sussman, J.L, Zaccai, G, Silman, I, Weik, M. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Substrate Traffic and Inhibition in Acetylcholinesterase.
Embo J., 25, 2006
|
|
5KIR
 
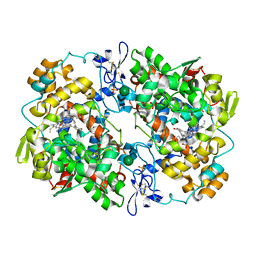 | | The Structure of Vioxx Bound to Human COX-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, GLYCEROL, ... | | Authors: | Orlando, B.J, Malkowski, M.G. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Crystal structure of rofecoxib bound to human cyclooxygenase-2.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3QJ4
 
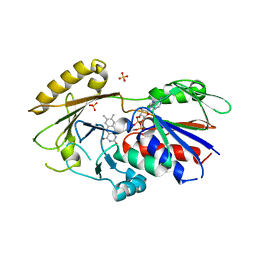 | | Crystal structure of Human Renalase (isoform 1) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Renalase, SULFATE ION | | Authors: | Milani, M, Ciriello, F, Baroni, S, Pandini, V, Aliverti, A, Canevari, G, Bolognesi, M. | | Deposit date: | 2011-01-28 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | FAD-binding site and NADP reactivity in human renalase: a new enzyme involved in blood pressure regulation
J.Mol.Biol., 411, 2011
|
|
5KOW
 
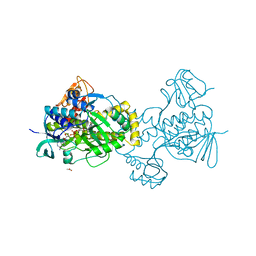 | | Structure of rifampicin monooxygenase | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pentachlorophenol 4-monooxygenase | | Authors: | Tanner, J.J, Liu, L.-K. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Antibiotic Deactivating, N-hydroxylating Rifampicin Monooxygenase.
J.Biol.Chem., 291, 2016
|
|
5KKC
 
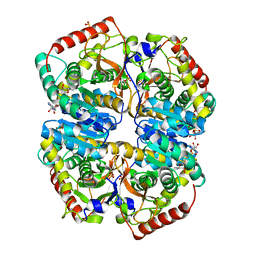 | | l-lactate dehydrogenase from rabbit muscle with the inhibitor 6DHNAD | | Descriptor: | L-lactate dehydrogenase A chain, SULFATE ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(5-aminocarbonyl-2~{H}-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Meneely, K.M, Moran, G.R, Lamb, A.L. | | Deposit date: | 2016-06-21 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Ligand binding phenomena that pertain to the metabolic function of renalase.
Arch.Biochem.Biophys., 612, 2016
|
|
7XTJ
 
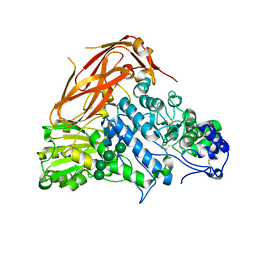 | | Crystal structure of E88A mutant of GH3 beta-xylosidase from Aspergillus niger (AnBX) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Kaenying, W, Kongsaeree, P.T, Tagami, T. | | Deposit date: | 2022-05-17 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and identification of amino acid residues for catalysis and binding of GH3 AnBX beta-xylosidase from Aspergillus niger.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
5KLS
 
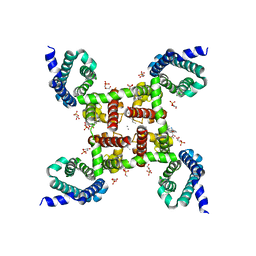 | | Structure of CavAb in complex with Br-dihydropyridine derivative UK-59811 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
5KPM
 
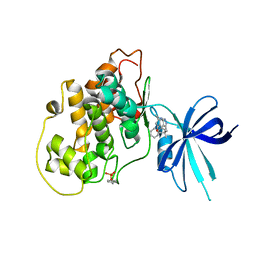 | | Glycogen Synthase Kinase 3 beta Complexed with BRD3731 | | Descriptor: | (4~{S})-3-(2,2-dimethylpropyl)-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KMH
 
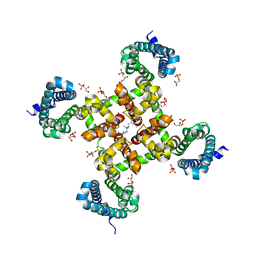 | | Structure of CavAb in complex with Br-verapamil | | Descriptor: | (2~{R})-2-(2-bromophenyl)-5-[2-(3,4-dimethoxyphenyl)ethyl-methyl-amino]-2-propan-2-yl-pentanenitrile, 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
5KN9
 
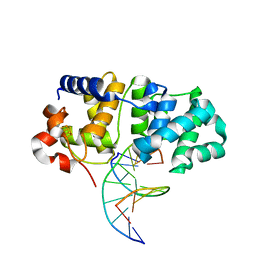 | | MutY N-terminal domain in complex with DNA containing an intrahelical oxoG:A base-pair | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*CP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Wang, L, Chakravarthy, S, Verdine, G.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
J. Biol. Chem., 292, 2017
|
|
5KML
 
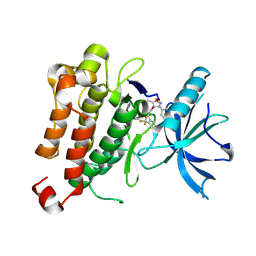 | |
5KPL
 
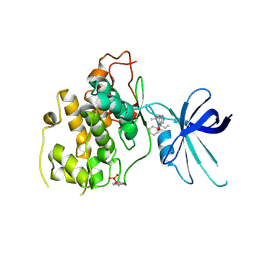 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0705 | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KRE
 
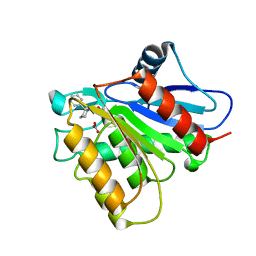 | | Covalent inhibitor of LYPLAL1 | | Descriptor: | (2~{R})-2-phenylpiperidine-1-carbaldehyde, Lysophospholipase-like protein 1, NITRATE ION | | Authors: | Pandit, J. | | Deposit date: | 2016-07-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a Selective Covalent Inhibitor of Lysophospholipase-like 1 (LYPLAL1) as a Tool to Evaluate the Role of this Serine Hydrolase in Metabolism.
Acs Chem.Biol., 11, 2016
|
|
5KVC
 
 | |
7XWM
 
 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
5KXK
 
 | | Hen Egg White Lysozyme at 100K, Data set 1 | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KXP
 
 | | Hen Egg White Lysozyme at 278K, Data set 2 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KXY
 
 | | Hen Egg White Lysozyme at 278K, Data set 8 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
