2J3L
 
 | | Prolyl-tRNA synthetase from Enterococcus faecalis complexed with a prolyl-adenylate analogue ('5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE) | | Descriptor: | '5'-O-(N-(L-PROLYL)-SULFAMOYL)ADENOSINE, PROLYL-TRNA SYNTHETASE, SULFATE ION | | Authors: | Crepin, T, Yaremchuk, A, Tukalo, M, Cusack, S. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Two Bacterial Prolyl-tRNA Synthetases with and without a Cis-Editing Domain.
Structure, 14, 2006
|
|
4JNA
 
 | | Crystal structure of the DepH complex with dimethyl-FK228 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DepH, ... | | Authors: | Li, J, Wang, C, Zhang, Z.M, Zhou, J.H, Cheng, E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of an NADP+-independent dithiol oxidase in FK228 biosynthesis.
Sci Rep, 4, 2014
|
|
1AA9
 
 | | HUMAN C-HA-RAS(1-171)(DOT)GDP, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | C-HA-RAS, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Ito, Y, Yamasaki, Y, Muto, Y, Kawai, G, Nishimura, S, Miyazawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1997-01-27 | | Release date: | 1997-07-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein.
Biochemistry, 36, 1997
|
|
4MZZ
 
 | |
4K24
 
 | | Structure of anti-uPAR Fab ATN-658 in complex with uPAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, ... | | Authors: | Huang, M.D, Xu, X, Yuan, C. | | Deposit date: | 2013-04-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Identification of a New Epitope in uPAR as a Target for the Cancer Therapeutic Monoclonal Antibody ATN-658, a Structural Homolog of the uPAR Binding Integrin CD11b ( alpha M)
Plos One, 9, 2014
|
|
2Z3Y
 
 | | Crystal structure of Lysine-specific demethylase1 | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL (2R,3S,4S)-5-[7,8-DIMETHYL-2,4-DIOXO-5-(3-PHENYLPROPANOYL)-1,3,4,5-TETRAHYDROBENZO[G]PTERIDIN-10(2H)-YL]-2,3,4-TRIHYDROXYPENTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Mimasu, S, Sengoku, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-06-08 | | Release date: | 2008-01-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25A
Biochem.Biophys.Res.Commun., 366, 2008
|
|
4Q05
 
 | | Crystal structure of an esterase E25 | | Descriptor: | SODIUM ION, esterase E25 | | Authors: | Li, P.Y, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for dimerization and catalysis of a novel esterase from the GTSAG motif subfamily of the bacterial hormone-sensitive lipase family
J.Biol.Chem., 289, 2014
|
|
2Z5U
 
 | |
1AEC
 
 | | CRYSTAL STRUCTURE OF ACTINIDIN-E-64 COMPLEX+ | | Descriptor: | ACTINIDIN, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Varughese, K.I. | | Deposit date: | 1992-02-05 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of an actinidin-E-64 complex.
Biochemistry, 31, 1992
|
|
4OUI
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with TRIACETYLCHITOTRIOSE (CTO) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2014-02-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1B41
 
 | | HUMAN ACETYLCHOLINESTERASE COMPLEXED WITH FASCICULIN-II, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Kryger, G, Harel, M, Shafferman, A, Silman, I, Sussman, J.L. | | Deposit date: | 1999-01-05 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of recombinant native and E202Q mutant human acetylcholinesterase complexed with the snake-venom toxin fasciculin-II.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4KRD
 
 | | Crystal Structure of Pho85-Pcl10 Complex | | Descriptor: | Cyclin-dependent protein kinase PHO85, PHO85 cyclin-10 | | Authors: | Quiocho, F.A, Zheng, F. | | Deposit date: | 2013-05-16 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | New Structural Insights into Phosphorylation-free Mechanism for Full Cyclin-dependent Kinase (CDK)-Cyclin Activity and Substrate Recognition.
J.Biol.Chem., 288, 2013
|
|
4O7J
 
 | |
4K23
 
 | | Structure of anti-uPAR Fab ATN-658 | | Descriptor: | anti-uPAR antibody, heavy chain, light chain | | Authors: | Yuan, C, Huang, M, Chen, L. | | Deposit date: | 2013-04-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a New Epitope in uPAR as a Target for the Cancer Therapeutic Monoclonal Antibody ATN-658, a Structural Homolog of the uPAR Binding Integrin CD11b ( alpha M)
Plos One, 9, 2014
|
|
1M5B
 
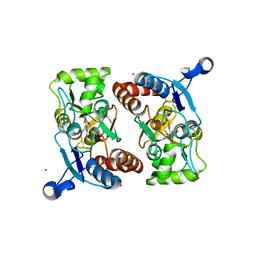 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
4K2F
 
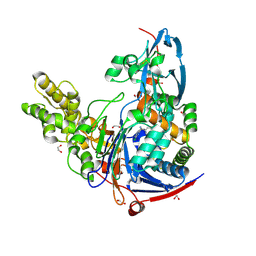 | | Structure of Pseudomonas aeruginosa PvdQ bound to BRD-A08522488 | | Descriptor: | (2S)-(4-chlorophenyl)(6-chloropyridin-2-yl)ethanenitrile, 1,2-ETHANEDIOL, Acyl-homoserine lactone acylase PvdQ | | Authors: | Drake, E.J, Wurst, J.M, Theriault, J.R, Munoz, B, Gulick, A.M. | | Deposit date: | 2013-04-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine.
Acs Chem.Biol., 9, 2014
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
4NYU
 
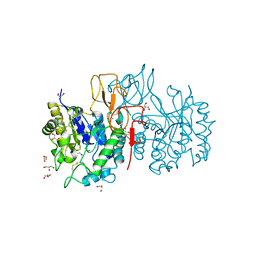 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in C 2 2 21 | | Descriptor: | ACETATE ION, CALCIUM ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4M1B
 
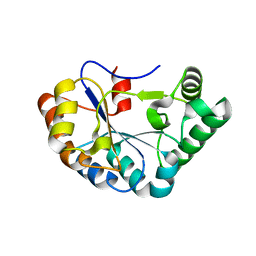 | |
4F08
 
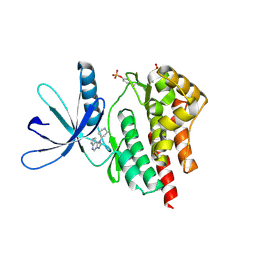 | |
4R50
 
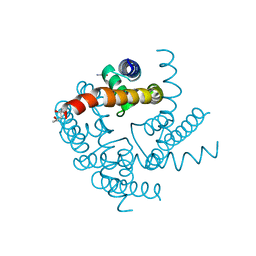 | | Crystal Structure of CNG mimicking NaK-ETPP mutant cocrystallized with Li+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, Potassium channel protein | | Authors: | De March, M, Napolitano, L.M.R, Onesti, S. | | Deposit date: | 2014-08-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2I4L
 
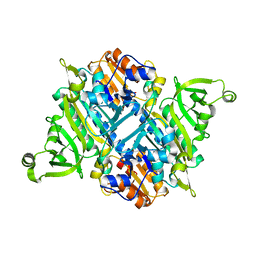 | |
4NY2
 
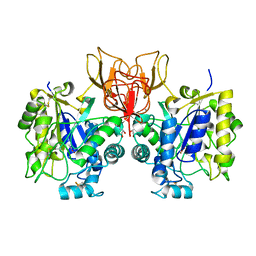 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in P 21 | | Descriptor: | ACETATE ION, CALCIUM ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NTJ
 
 | | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12, ... | | Authors: | Zhang, K, Zhang, J, Gao, Z.-G, Zhang, D, Zhu, L, Han, G.W, Moss, S.M, Paoletta, S, Kiselev, E, Lu, W, Fenalti, G, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the human P2Y12 receptor in complex with an antithrombotic drug
Nature, 509, 2014
|
|
1VWK
 
 | | STREPTAVIDIN-CYCLO-[5-S-VALERAMIDE-HPQGPPC]K-NH2 | | Descriptor: | PENTANOIC ACID, PEPTIDE LIGAND CONTAINING HPQ, STREPTAVIDIN | | Authors: | Katz, B.A, Cass, R.T. | | Deposit date: | 1997-03-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | In crystals of complexes of streptavidin with peptide ligands containing the HPQ sequence the pKa of the peptide histidine is less than 3.0.
J.Biol.Chem., 272, 1997
|
|
